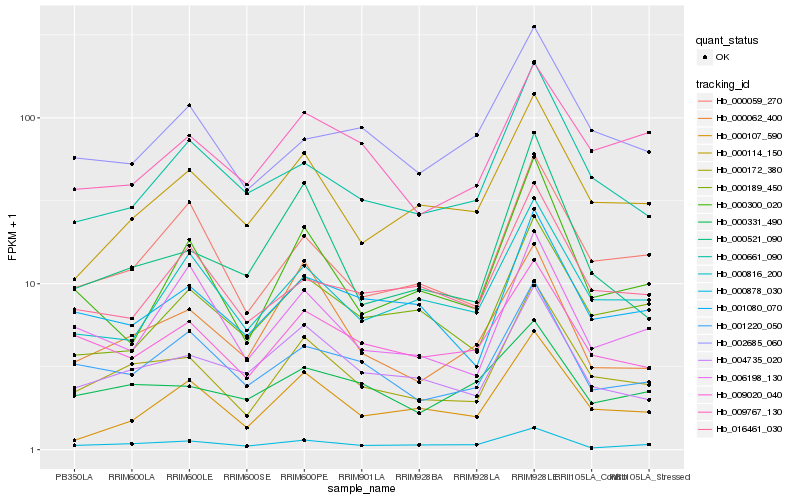
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000172_380 |
0.0 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 2 |
Hb_000059_270 |
0.1161380645 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 3 |
Hb_004735_020 |
0.1225036599 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 4 |
Hb_001080_070 |
0.1290405149 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 5 |
Hb_000114_150 |
0.12918057 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 6 |
Hb_001220_050 |
0.1304483319 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 7 |
Hb_009020_040 |
0.1309994662 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000661_090 |
0.1338712281 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000521_090 |
0.1345442811 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000878_030 |
0.1356044475 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 11 |
Hb_000300_020 |
0.1390443043 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000062_400 |
0.1393292077 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000189_450 |
0.1393998283 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_000107_590 |
0.141534347 |
- |
- |
PREDICTED: tricalbin-3 [Jatropha curcas] |
| 15 |
Hb_000816_200 |
0.1434002991 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 16 |
Hb_009767_130 |
0.1442809222 |
- |
- |
PREDICTED: preprotein translocase subunit SECE1 [Jatropha curcas] |
| 17 |
Hb_016461_030 |
0.1457089235 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 18 |
Hb_006198_130 |
0.1457321995 |
- |
- |
PREDICTED: uncharacterized protein LOC105644406 [Jatropha curcas] |
| 19 |
Hb_002685_060 |
0.1494323152 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 20 |
Hb_000331_490 |
0.1503988266 |
- |
- |
PREDICTED: radical S-adenosyl methionine domain-containing protein 1, mitochondrial [Jatropha curcas] |