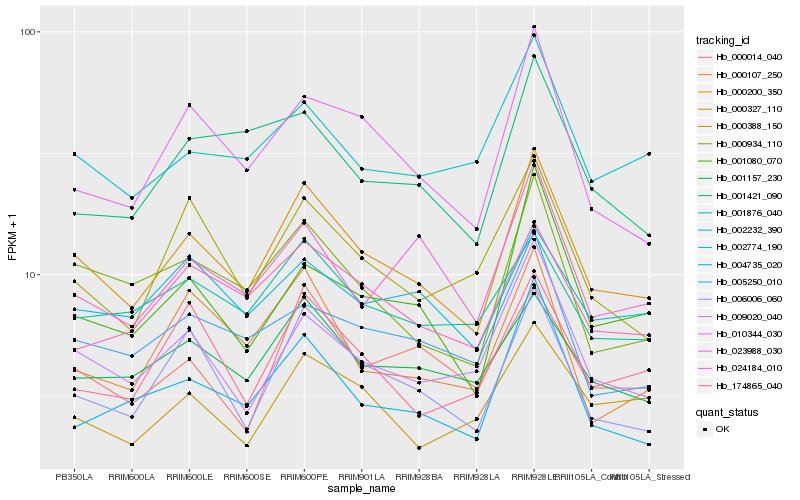
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000200_350 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636929 [Jatropha curcas] |
| 2 |
Hb_006006_060 |
0.0851855916 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000934_110 |
0.0923438529 |
- |
- |
PREDICTED: protein trichome berefringence-like 7 [Jatropha curcas] |
| 4 |
Hb_010344_030 |
0.0942131776 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_009020_040 |
0.0953792598 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001157_230 |
0.0961982911 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 7 |
Hb_024184_010 |
0.1026728748 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 8 |
Hb_005250_010 |
0.1035148886 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 9 |
Hb_001080_070 |
0.10356913 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 10 |
Hb_174865_040 |
0.1051492524 |
- |
- |
PREDICTED: crt homolog 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004735_020 |
0.105794812 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 12 |
Hb_001876_040 |
0.105872956 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 13 |
Hb_000388_150 |
0.1066687014 |
- |
- |
hypothetical protein JCGZ_21651 [Jatropha curcas] |
| 14 |
Hb_002232_390 |
0.1076504857 |
- |
- |
PREDICTED: uncharacterized protein LOC105635994 [Jatropha curcas] |
| 15 |
Hb_000014_040 |
0.1078873041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000327_110 |
0.1114848385 |
- |
- |
hypothetical protein JCGZ_06657 [Jatropha curcas] |
| 17 |
Hb_000107_250 |
0.1124007534 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 18 |
Hb_023988_030 |
0.1129954228 |
- |
- |
PREDICTED: transmembrane protein 19 [Vitis vinifera] |
| 19 |
Hb_001421_090 |
0.1137751901 |
- |
- |
hypothetical protein RCOM_1278080 [Ricinus communis] |
| 20 |
Hb_002774_190 |
0.1139770765 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |