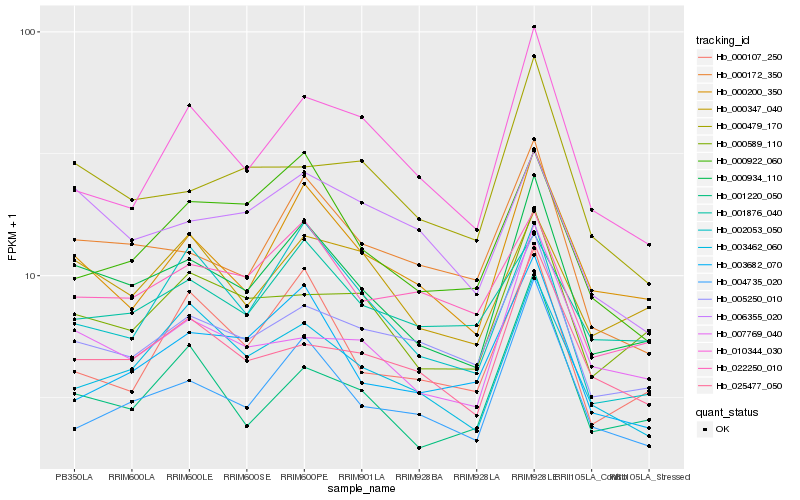
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000934_110 |
0.0 |
- |
- |
PREDICTED: protein trichome berefringence-like 7 [Jatropha curcas] |
| 2 |
Hb_000200_350 |
0.0923438529 |
- |
- |
PREDICTED: uncharacterized protein LOC105636929 [Jatropha curcas] |
| 3 |
Hb_000172_350 |
0.0997262112 |
- |
- |
PREDICTED: uncharacterized protein LOC105650963 [Jatropha curcas] |
| 4 |
Hb_000589_110 |
0.1031994539 |
- |
- |
PREDICTED: uncharacterized protein LOC105647614 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005250_010 |
0.1047547199 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 6 |
Hb_007769_040 |
0.1084693898 |
- |
- |
PREDICTED: probable threonine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 7 |
Hb_003462_060 |
0.1108893691 |
- |
- |
PREDICTED: ABC transporter G family member 24-like [Jatropha curcas] |
| 8 |
Hb_000347_040 |
0.1110738241 |
- |
- |
hypothetical protein JCGZ_24660 [Jatropha curcas] |
| 9 |
Hb_025477_050 |
0.1119825071 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_004735_020 |
0.1125269546 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 11 |
Hb_000479_170 |
0.1166007856 |
- |
- |
chaperone clpb, putative [Ricinus communis] |
| 12 |
Hb_022250_010 |
0.1168744025 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 13 |
Hb_002053_050 |
0.1170529182 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 14 |
Hb_000107_250 |
0.1188945787 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 15 |
Hb_001220_050 |
0.1193148693 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 16 |
Hb_010344_030 |
0.1217421783 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001876_040 |
0.1219155874 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 18 |
Hb_003682_070 |
0.1224630817 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006355_020 |
0.1226706319 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 20 |
Hb_000922_060 |
0.1249659117 |
- |
- |
kinesin light chain, putative [Ricinus communis] |