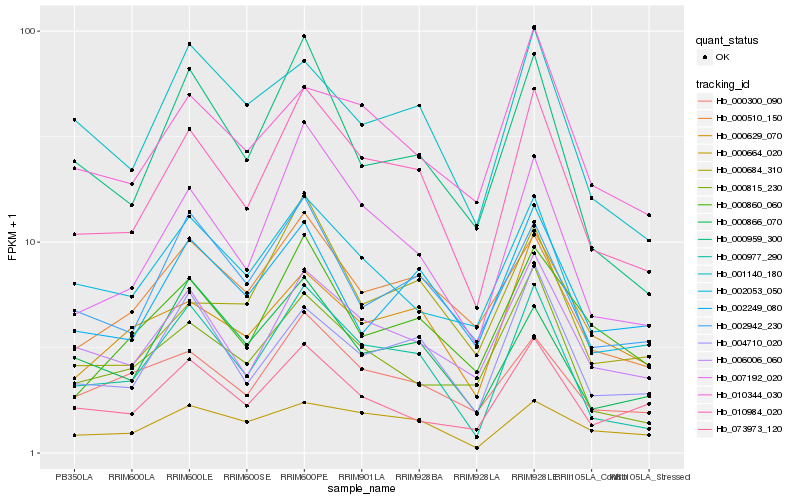
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010984_020 |
0.0 |
- |
- |
Translation factor GUF1 like, chloroplastic [Glycine soja] |
| 2 |
Hb_000977_290 |
0.07680546 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_007192_020 |
0.0817477238 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |
| 4 |
Hb_006006_060 |
0.0976451243 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004710_020 |
0.1111695907 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 6 |
Hb_000959_300 |
0.1141420626 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 7 |
Hb_000510_150 |
0.1161343925 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000300_090 |
0.1166238276 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_002053_050 |
0.1187675371 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 10 |
Hb_002942_230 |
0.1209966578 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 11 |
Hb_000664_020 |
0.1233064974 |
- |
- |
tryptophanyl-tRNA synthetase, putative [Ricinus communis] |
| 12 |
Hb_000860_060 |
0.1279907016 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 [Jatropha curcas] |
| 13 |
Hb_010344_030 |
0.1282694659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001140_180 |
0.1303524718 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 15 |
Hb_000629_070 |
0.1304228828 |
- |
- |
DNA-3-methyladenine glycosylase, putative [Ricinus communis] |
| 16 |
Hb_002249_080 |
0.1311111378 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 17 |
Hb_000815_230 |
0.1314570167 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_073973_120 |
0.1331561523 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_000866_070 |
0.1340128173 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SKP1-like protein 21 [Jatropha curcas] |
| 20 |
Hb_000684_310 |
0.1353036162 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |