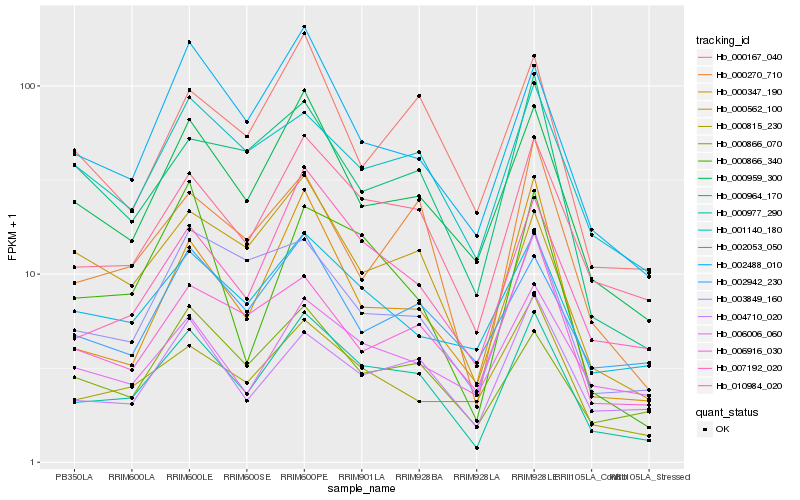
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000977_290 |
0.0 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_010984_020 |
0.07680546 |
- |
- |
Translation factor GUF1 like, chloroplastic [Glycine soja] |
| 3 |
Hb_000959_300 |
0.1050434745 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 4 |
Hb_007192_020 |
0.1096640356 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |
| 5 |
Hb_002488_010 |
0.1275806786 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 6 |
Hb_004710_020 |
0.1279646249 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 7 |
Hb_001140_180 |
0.1314643799 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 8 |
Hb_000866_070 |
0.1315933724 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SKP1-like protein 21 [Jatropha curcas] |
| 9 |
Hb_000562_100 |
0.1325052845 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 10 |
Hb_000270_710 |
0.1334400692 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_002053_050 |
0.1346615515 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 12 |
Hb_006006_060 |
0.1419487412 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000866_340 |
0.1422963789 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_002942_230 |
0.1427175255 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 15 |
Hb_006916_030 |
0.1429879553 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 16 |
Hb_000815_230 |
0.1430780362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003849_160 |
0.1433952185 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 18 |
Hb_000167_040 |
0.1437042509 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 19 |
Hb_000964_170 |
0.1447722011 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |
| 20 |
Hb_000347_190 |
0.1475868569 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |