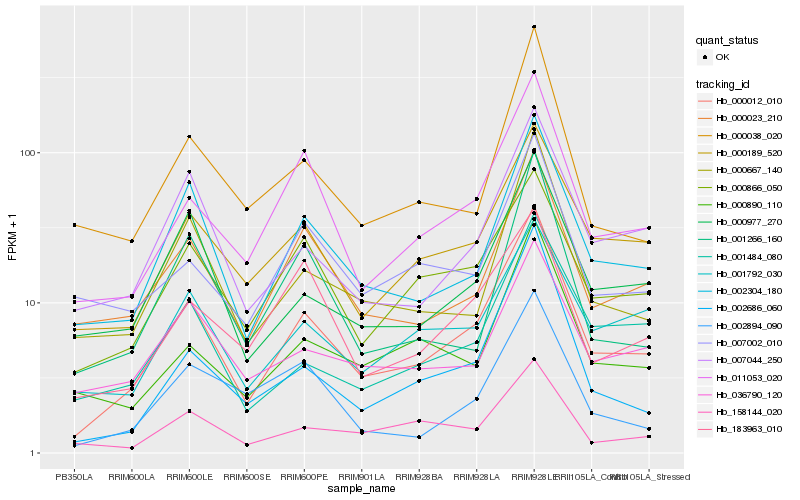
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000012_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_07709 [Jatropha curcas] |
| 2 |
Hb_002304_180 |
0.1146836611 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlI, chloroplastic [Jatropha curcas] |
| 3 |
Hb_011053_020 |
0.1186790858 |
- |
- |
lipoic acid synthetase, putative [Ricinus communis] |
| 4 |
Hb_000189_520 |
0.1272043553 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 5 |
Hb_000023_210 |
0.1338990046 |
- |
- |
PREDICTED: protein TIC 55, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000667_140 |
0.1381965131 |
- |
- |
PREDICTED: uridine kinase-like protein 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000866_050 |
0.1388172584 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 8 |
Hb_007044_250 |
0.1394038516 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002686_060 |
0.1412526934 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000977_270 |
0.1439385315 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001484_080 |
0.1440522302 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 12 |
Hb_001792_030 |
0.1494552234 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 13 |
Hb_000890_110 |
0.1521731366 |
- |
- |
PREDICTED: GTP-binding protein OBGC, chloroplastic [Populus euphratica] |
| 14 |
Hb_000038_020 |
0.1556404696 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001266_160 |
0.1570481632 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_158144_020 |
0.1578963904 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Vitis vinifera] |
| 17 |
Hb_183963_010 |
0.1589304907 |
- |
- |
core region of GTP cyclohydrolase I family protein [Populus trichocarpa] |
| 18 |
Hb_007002_010 |
0.1590029227 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 19 |
Hb_036790_120 |
0.1605655203 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002894_090 |
0.162373753 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase [Jatropha curcas] |