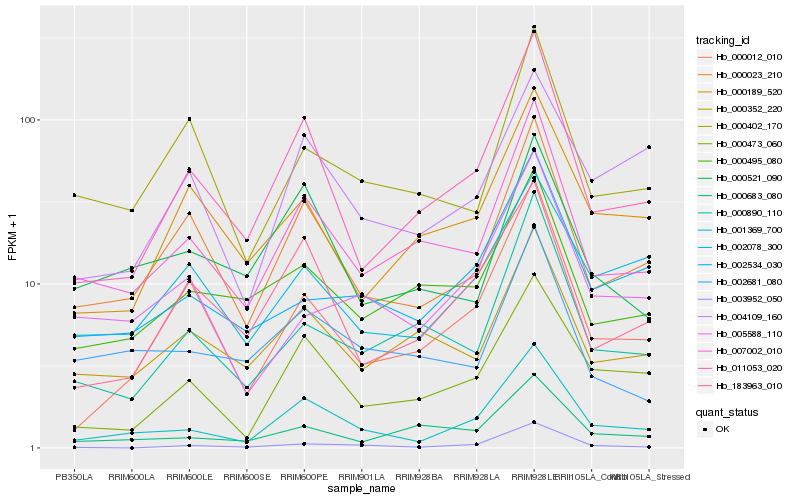
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_700 |
0.0 |
- |
- |
- |
| 2 |
Hb_011053_020 |
0.1359890749 |
- |
- |
lipoic acid synthetase, putative [Ricinus communis] |
| 3 |
Hb_000473_060 |
0.1401286353 |
- |
- |
PREDICTED: psbP domain-containing protein 2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000023_210 |
0.1483775282 |
- |
- |
PREDICTED: protein TIC 55, chloroplastic [Jatropha curcas] |
| 5 |
Hb_007002_010 |
0.1519558054 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 6 |
Hb_183963_010 |
0.1690059562 |
- |
- |
core region of GTP cyclohydrolase I family protein [Populus trichocarpa] |
| 7 |
Hb_000012_010 |
0.1698336991 |
- |
- |
hypothetical protein JCGZ_07709 [Jatropha curcas] |
| 8 |
Hb_002534_030 |
0.1814598439 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 9 |
Hb_004109_160 |
0.1825012136 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_003952_050 |
0.1878743025 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 11 |
Hb_005588_110 |
0.190469182 |
- |
- |
hypothetical protein POPTR_0002s12860g [Populus trichocarpa] |
| 12 |
Hb_002078_300 |
0.1905806651 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000683_080 |
0.193036503 |
- |
- |
hypothetical protein EUGRSUZ_G013602, partial [Eucalyptus grandis] |
| 14 |
Hb_000890_110 |
0.1940812359 |
- |
- |
PREDICTED: GTP-binding protein OBGC, chloroplastic [Populus euphratica] |
| 15 |
Hb_000521_090 |
0.1942856956 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000495_080 |
0.1971215652 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 17 |
Hb_002681_080 |
0.1973302504 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000352_220 |
0.1984590599 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000189_520 |
0.1988737913 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 20 |
Hb_000402_170 |
0.202893435 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |