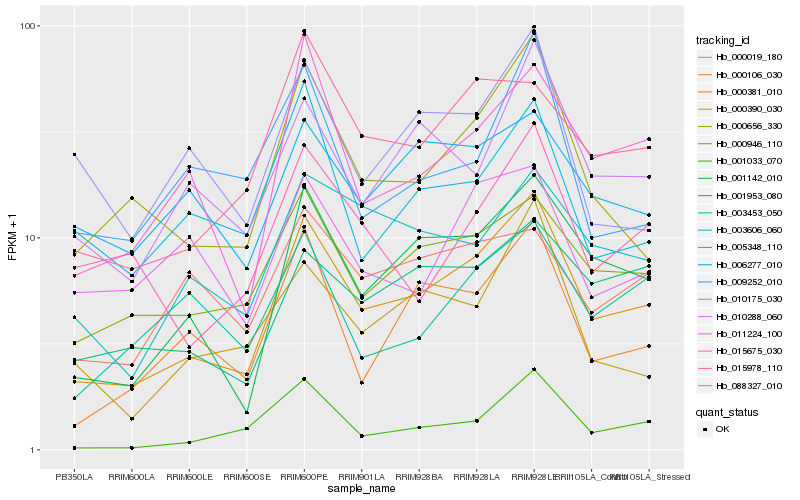
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000381_010 |
0.0 |
- |
- |
PREDICTED: arogenate dehydratase/prephenate dehydratase 1, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_001142_010 |
0.1084626654 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 6 [Jatropha curcas] |
| 3 |
Hb_000656_330 |
0.1298786048 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 4 |
Hb_000946_110 |
0.1329947556 |
- |
- |
GTP cyclohydrolase I, putative [Ricinus communis] |
| 5 |
Hb_015675_030 |
0.1426576971 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000106_030 |
0.1432041345 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_010288_060 |
0.1444648644 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 8 |
Hb_088327_010 |
0.1500351511 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 9 |
Hb_001953_080 |
0.1528019588 |
- |
- |
pseudouridine synthase family protein, partial [Manihot esculenta] |
| 10 |
Hb_000390_030 |
0.1561699429 |
- |
- |
hypothetical protein ZEAMMB73_364791 [Zea mays] |
| 11 |
Hb_003606_060 |
0.1565641438 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 12 |
Hb_001033_070 |
0.1571615532 |
- |
- |
seryl-tRNA synthetase, putative [Ricinus communis] |
| 13 |
Hb_015978_110 |
0.1664392733 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g62470, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_005348_110 |
0.1675324223 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 15 |
Hb_006277_010 |
0.1705320766 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Gossypium raimondii] |
| 16 |
Hb_003453_050 |
0.1714077239 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 17 |
Hb_000019_180 |
0.1734329136 |
transcription factor |
TF Family: SET |
Histone-lysine N-methyltransferase family protein [Populus trichocarpa] |
| 18 |
Hb_010175_030 |
0.1735712226 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 19 |
Hb_011224_100 |
0.1755979802 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_009252_010 |
0.1763945642 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |