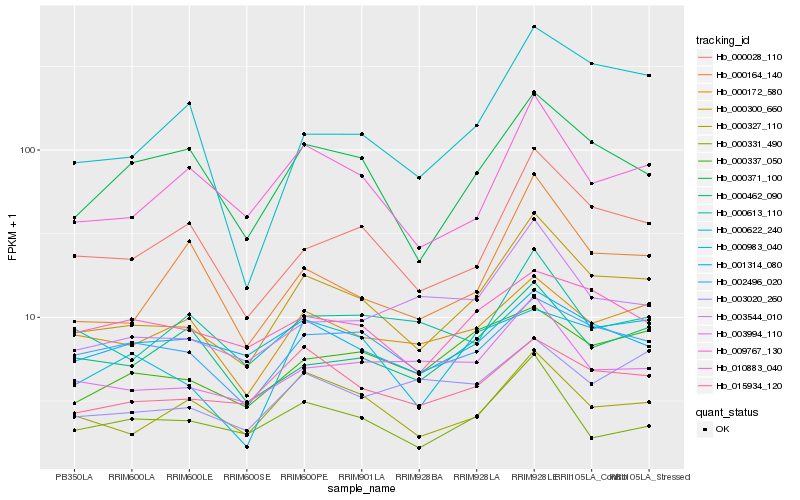
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_660 |
0.0 |
- |
- |
unknown [Glycine max] |
| 2 |
Hb_000028_110 |
0.11617631 |
- |
- |
PREDICTED: uncharacterized protein LOC105645305 [Jatropha curcas] |
| 3 |
Hb_009767_130 |
0.1188132833 |
- |
- |
PREDICTED: preprotein translocase subunit SECE1 [Jatropha curcas] |
| 4 |
Hb_000371_100 |
0.1250706229 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
RecName: Full=Geranylgeranyl pyrophosphate synthase, chloroplastic; Short=GGPP synthase; Short=GGPS; AltName: Full=(2E,6E)-farnesyl diphosphate synthase; AltName: Full=Dimethylallyltranstransferase; AltName: Full=Farnesyl diphosphate synthase; AltName: Full=Farnesyltranstransferase; AltName: Full=Geranyltranstransferase; Flags: Precursor [Hevea brasiliensis] |
| 5 |
Hb_015934_120 |
0.127174303 |
- |
- |
PREDICTED: callose synthase 7 [Vitis vinifera] |
| 6 |
Hb_000327_110 |
0.127237276 |
- |
- |
hypothetical protein JCGZ_06657 [Jatropha curcas] |
| 7 |
Hb_003994_110 |
0.1324492852 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646130 [Jatropha curcas] |
| 8 |
Hb_000337_050 |
0.1325474558 |
- |
- |
PREDICTED: chloroplast envelope membrane protein [Jatropha curcas] |
| 9 |
Hb_002496_020 |
0.1346460821 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_003544_010 |
0.1348583596 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit A, chloroplastic/mitochondrial [Jatropha curcas] |
| 11 |
Hb_000164_140 |
0.1365067718 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 12 |
Hb_000172_580 |
0.1391814221 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000331_490 |
0.1424699436 |
- |
- |
PREDICTED: radical S-adenosyl methionine domain-containing protein 1, mitochondrial [Jatropha curcas] |
| 14 |
Hb_010883_040 |
0.1429733306 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 15 |
Hb_000462_090 |
0.1451895289 |
- |
- |
PREDICTED: carotene epsilon-monooxygenase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003020_260 |
0.1463783685 |
- |
- |
PREDICTED: metal tolerance protein B [Jatropha curcas] |
| 17 |
Hb_000983_040 |
0.1465326686 |
- |
- |
PREDICTED: transmembrane protein 136 [Jatropha curcas] |
| 18 |
Hb_001314_080 |
0.1470475663 |
- |
- |
hypothetical protein POPTR_0018s14330g [Populus trichocarpa] |
| 19 |
Hb_000622_240 |
0.1472307877 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000613_110 |
0.1473957557 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |