| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
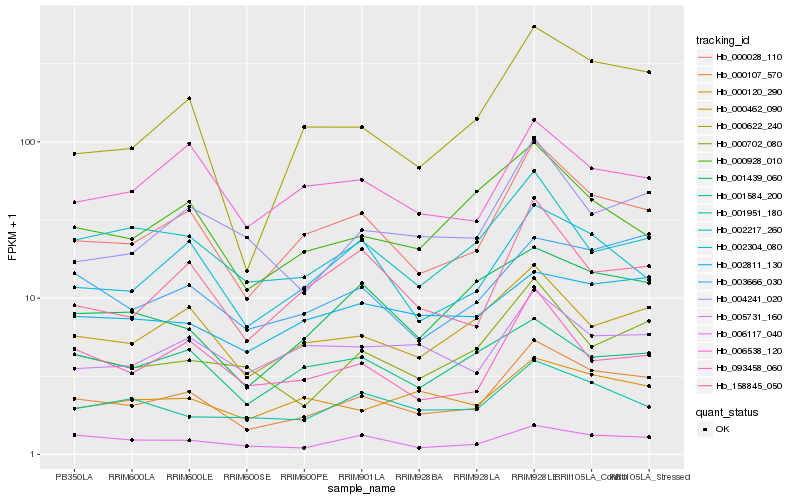
Hb_000107_570 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634044 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000028_110 |
0.0741023848 |
- |
- |
PREDICTED: uncharacterized protein LOC105645305 [Jatropha curcas] |
| 3 |
Hb_000622_240 |
0.1138766009 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000462_090 |
0.1146372258 |
- |
- |
PREDICTED: carotene epsilon-monooxygenase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003666_030 |
0.1190652347 |
- |
- |
Thiol:disulfide interchange protein txlA, putative [Ricinus communis] |
| 6 |
Hb_000702_080 |
0.1246059097 |
- |
- |
PREDICTED: uncharacterized protein LOC105630492 [Jatropha curcas] |
| 7 |
Hb_006117_040 |
0.1252189028 |
- |
- |
PREDICTED: seed biotin-containing protein SBP65 [Jatropha curcas] |
| 8 |
Hb_002304_080 |
0.1265539244 |
- |
- |
PREDICTED: uncharacterized protein LOC105649626 [Jatropha curcas] |
| 9 |
Hb_093458_060 |
0.1290261163 |
- |
- |
PREDICTED: uncharacterized protein LOC105633194 [Jatropha curcas] |
| 10 |
Hb_158845_050 |
0.1297482011 |
- |
- |
PREDICTED: uncharacterized protein LOC105122890 [Populus euphratica] |
| 11 |
Hb_001439_060 |
0.1304403652 |
- |
- |
PREDICTED: uncharacterized protein LOC105644108 [Jatropha curcas] |
| 12 |
Hb_004241_020 |
0.1311931181 |
- |
- |
PREDICTED: DAG protein, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_005731_160 |
0.1335204792 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_001951_180 |
0.1339913167 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_000120_290 |
0.1345428997 |
- |
- |
PREDICTED: N-acylphosphatidylethanolamine synthase isoform X1 [Jatropha curcas] |
| 16 |
Hb_006538_120 |
0.1346005639 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002811_130 |
0.1367907368 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_002217_260 |
0.138962126 |
- |
- |
PREDICTED: uncharacterized protein LOC105643577 [Jatropha curcas] |
| 19 |
Hb_001584_200 |
0.1394318128 |
- |
- |
PREDICTED: Golgi apparatus membrane protein-like protein ECHIDNA [Jatropha curcas] |
| 20 |
Hb_000928_010 |
0.1406295474 |
- |
- |
Cell division protein ftsZ, putative [Ricinus communis] |