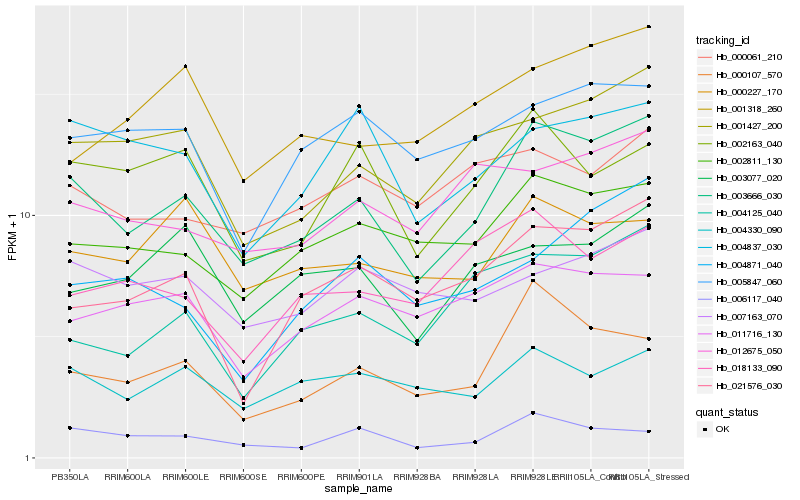
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003666_030 |
0.0 |
- |
- |
Thiol:disulfide interchange protein txlA, putative [Ricinus communis] |
| 2 |
Hb_001427_200 |
0.1157114792 |
- |
- |
PREDICTED: uncharacterized protein LOC105637796 [Jatropha curcas] |
| 3 |
Hb_002811_130 |
0.1175845809 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_000107_570 |
0.1190652347 |
- |
- |
PREDICTED: uncharacterized protein LOC105634044 isoform X1 [Jatropha curcas] |
| 5 |
Hb_021576_030 |
0.1286180424 |
- |
- |
PREDICTED: uncharacterized protein LOC105637542 [Jatropha curcas] |
| 6 |
Hb_006117_040 |
0.1289377899 |
- |
- |
PREDICTED: seed biotin-containing protein SBP65 [Jatropha curcas] |
| 7 |
Hb_012675_050 |
0.1322147277 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004330_090 |
0.1341908698 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_004125_040 |
0.1350474352 |
- |
- |
- |
| 10 |
Hb_004837_030 |
0.1385841156 |
- |
- |
PREDICTED: mitochondrial tRNA-specific 2-thiouridylase 1 [Jatropha curcas] |
| 11 |
Hb_002163_040 |
0.1395938389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005847_060 |
0.1402528412 |
- |
- |
hypothetical protein POPTR_0006s02920g [Populus trichocarpa] |
| 13 |
Hb_003077_020 |
0.1409260332 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_001318_260 |
0.1424620188 |
- |
- |
PREDICTED: nitrogen regulatory protein P-II homolog [Jatropha curcas] |
| 15 |
Hb_000061_210 |
0.1427287717 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_018133_090 |
0.1444215861 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000227_170 |
0.1444758631 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 18 |
Hb_007163_070 |
0.1446660845 |
- |
- |
PREDICTED: tRNA-dihydrouridine(20) synthase [NAD(P)+]-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_011716_130 |
0.1468682952 |
- |
- |
PREDICTED: probable acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, mitochondrial isoform X5 [Populus euphratica] |
| 20 |
Hb_004871_040 |
0.1471405272 |
- |
- |
PREDICTED: tRNA (guanine-N(7)-)-methyltransferase [Jatropha curcas] |