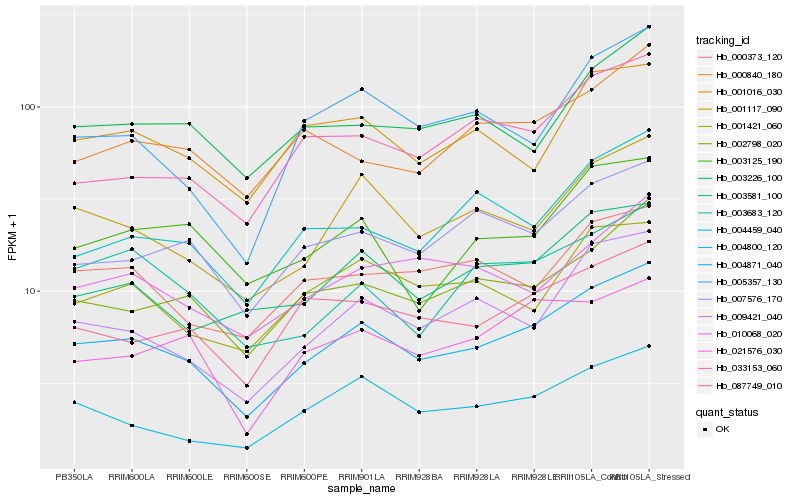
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004871_040 |
0.0 |
- |
- |
PREDICTED: tRNA (guanine-N(7)-)-methyltransferase [Jatropha curcas] |
| 2 |
Hb_001117_090 |
0.0863872094 |
- |
- |
PREDICTED: NADPH-dependent pterin aldehyde reductase [Jatropha curcas] |
| 3 |
Hb_002798_020 |
0.0901206887 |
- |
- |
PREDICTED: uncharacterized protein LOC105647735 [Jatropha curcas] |
| 4 |
Hb_009421_040 |
0.0906296107 |
- |
- |
PREDICTED: eukaryotic initiation factor 4A [Jatropha curcas] |
| 5 |
Hb_004800_120 |
0.0942630259 |
- |
- |
PREDICTED: maf-like protein DDB_G0281937 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003581_100 |
0.0953353881 |
- |
- |
PREDICTED: protein TWIN LOV 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_087749_010 |
0.0963167884 |
- |
- |
PREDICTED: uncharacterized protein LOC105631194 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007576_170 |
0.0985903274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005357_130 |
0.1043297293 |
- |
- |
hypothetical protein L484_012905 [Morus notabilis] |
| 10 |
Hb_021576_030 |
0.1043647612 |
- |
- |
PREDICTED: uncharacterized protein LOC105637542 [Jatropha curcas] |
| 11 |
Hb_004459_040 |
0.1049742261 |
- |
- |
PREDICTED: uncharacterized protein LOC105649649 [Jatropha curcas] |
| 12 |
Hb_001421_060 |
0.1066051767 |
- |
- |
PREDICTED: tobamovirus multiplication protein 2B isoform X2 [Jatropha curcas] |
| 13 |
Hb_033153_060 |
0.1066982532 |
- |
- |
Transcription elongation factor, putative [Ricinus communis] |
| 14 |
Hb_003125_190 |
0.1079250931 |
- |
- |
hypothetical protein POPTR_0015s10960g [Populus trichocarpa] |
| 15 |
Hb_000373_120 |
0.1081359482 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105629120 [Jatropha curcas] |
| 16 |
Hb_003683_120 |
0.1090826834 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001016_030 |
0.1107878223 |
- |
- |
PREDICTED: uncharacterized protein LOC105636529 isoform X2 [Jatropha curcas] |
| 18 |
Hb_003226_100 |
0.1111694363 |
- |
- |
PREDICTED: 60S ribosomal protein L18-2 [Jatropha curcas] |
| 19 |
Hb_010068_020 |
0.1112108134 |
- |
- |
PREDICTED: uncharacterized protein LOC105632966 [Jatropha curcas] |
| 20 |
Hb_000840_180 |
0.112177522 |
- |
- |
protein BOLA2 [Jatropha curcas] |