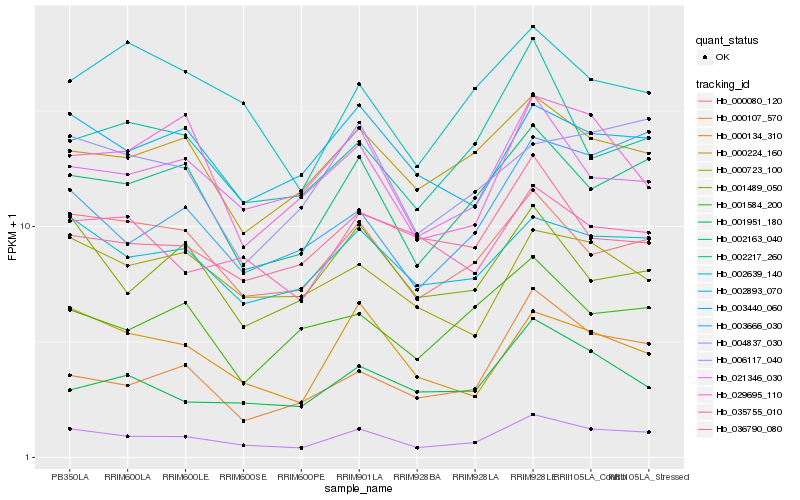
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006117_040 |
0.0 |
- |
- |
PREDICTED: seed biotin-containing protein SBP65 [Jatropha curcas] |
| 2 |
Hb_002163_040 |
0.0840402854 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000080_120 |
0.0918849206 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350-like [Jatropha curcas] |
| 4 |
Hb_002893_070 |
0.094323823 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_021346_030 |
0.1016021783 |
- |
- |
PREDICTED: uncharacterized protein LOC105141875 [Populus euphratica] |
| 6 |
Hb_000224_160 |
0.1048061701 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 7 |
Hb_000134_310 |
0.1067562691 |
- |
- |
PREDICTED: protein gamma response 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003440_060 |
0.1095014878 |
- |
- |
PREDICTED: translin-associated protein X isoform X1 [Jatropha curcas] |
| 9 |
Hb_000723_100 |
0.1169271175 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 10 |
Hb_001489_050 |
0.1185690853 |
- |
- |
PREDICTED: protein TWIN LOV 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002217_260 |
0.1189087047 |
- |
- |
PREDICTED: uncharacterized protein LOC105643577 [Jatropha curcas] |
| 12 |
Hb_001951_180 |
0.1205610881 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_004837_030 |
0.1217708434 |
- |
- |
PREDICTED: mitochondrial tRNA-specific 2-thiouridylase 1 [Jatropha curcas] |
| 14 |
Hb_029695_110 |
0.1229291595 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 15 |
Hb_002639_140 |
0.1238615443 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 5 [Jatropha curcas] |
| 16 |
Hb_035755_010 |
0.1247193542 |
- |
- |
unknown [Populus trichocarpa] |
| 17 |
Hb_001584_200 |
0.1249377299 |
- |
- |
PREDICTED: Golgi apparatus membrane protein-like protein ECHIDNA [Jatropha curcas] |
| 18 |
Hb_000107_570 |
0.1252189028 |
- |
- |
PREDICTED: uncharacterized protein LOC105634044 isoform X1 [Jatropha curcas] |
| 19 |
Hb_036790_080 |
0.1256067065 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 20 |
Hb_003666_030 |
0.1289377899 |
- |
- |
Thiol:disulfide interchange protein txlA, putative [Ricinus communis] |