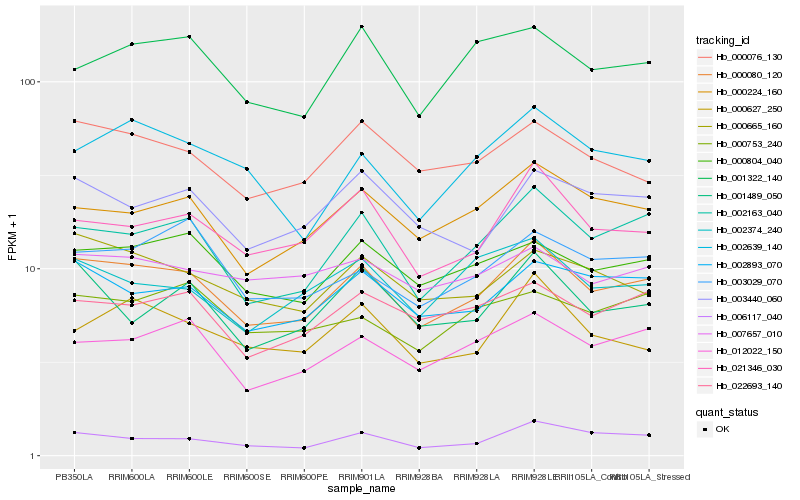
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000080_120 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350-like [Jatropha curcas] |
| 2 |
Hb_002163_040 |
0.0698685706 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002893_070 |
0.0751108155 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000804_040 |
0.0796056399 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_000224_160 |
0.0821124262 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 6 |
Hb_000076_130 |
0.0824459853 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 7 |
Hb_012022_150 |
0.0832366889 |
- |
- |
PREDICTED: protein root UVB sensitive 5 [Jatropha curcas] |
| 8 |
Hb_000665_160 |
0.0849408017 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X1 [Jatropha curcas] |
| 9 |
Hb_022693_140 |
0.0863114622 |
- |
- |
PREDICTED: uncharacterized protein LOC105632654 isoform X1 [Jatropha curcas] |
| 10 |
Hb_021346_030 |
0.0865985682 |
- |
- |
PREDICTED: uncharacterized protein LOC105141875 [Populus euphratica] |
| 11 |
Hb_003440_060 |
0.0884204431 |
- |
- |
PREDICTED: translin-associated protein X isoform X1 [Jatropha curcas] |
| 12 |
Hb_006117_040 |
0.0918849206 |
- |
- |
PREDICTED: seed biotin-containing protein SBP65 [Jatropha curcas] |
| 13 |
Hb_003029_070 |
0.0933618359 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_001322_140 |
0.0955635606 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001489_050 |
0.095718491 |
- |
- |
PREDICTED: protein TWIN LOV 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002639_140 |
0.0960234469 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 5 [Jatropha curcas] |
| 17 |
Hb_000627_250 |
0.0960354482 |
- |
- |
PREDICTED: uncharacterized protein LOC105632682 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002374_240 |
0.0968403177 |
- |
- |
nucellin, putative [Ricinus communis] |
| 19 |
Hb_000753_240 |
0.0971696675 |
- |
- |
PREDICTED: ADP,ATP carrier protein ER-ANT1 [Jatropha curcas] |
| 20 |
Hb_007657_010 |
0.097804833 |
- |
- |
ATP binding protein, putative [Ricinus communis] |