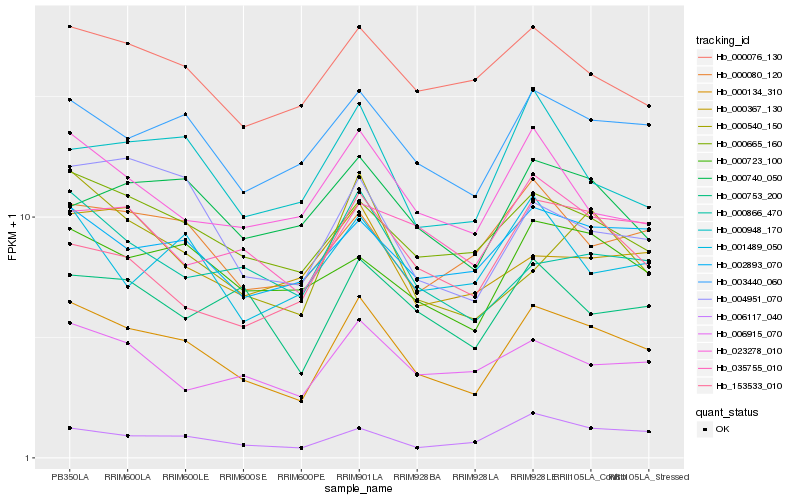
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000134_310 |
0.0 |
- |
- |
PREDICTED: protein gamma response 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003440_060 |
0.0960756056 |
- |
- |
PREDICTED: translin-associated protein X isoform X1 [Jatropha curcas] |
| 3 |
Hb_000665_160 |
0.1006470803 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X1 [Jatropha curcas] |
| 4 |
Hb_006117_040 |
0.1067562691 |
- |
- |
PREDICTED: seed biotin-containing protein SBP65 [Jatropha curcas] |
| 5 |
Hb_002893_070 |
0.1075010509 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000723_100 |
0.1138754961 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 7 |
Hb_000080_120 |
0.114199555 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350-like [Jatropha curcas] |
| 8 |
Hb_023278_010 |
0.1155558172 |
- |
- |
Chaperone protein DnaJ [Glycine soja] |
| 9 |
Hb_006915_070 |
0.1163504247 |
- |
- |
hypothetical protein CAPTEDRAFT_111329, partial [Capitella teleta] |
| 10 |
Hb_004951_070 |
0.1169820854 |
desease resistance |
Gene Name: AAA |
Stage III sporulation protein AA, putative [Ricinus communis] |
| 11 |
Hb_000076_130 |
0.117202275 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 12 |
Hb_000866_470 |
0.1199301751 |
- |
- |
PREDICTED: uncharacterized protein LOC105643015 [Jatropha curcas] |
| 13 |
Hb_000948_170 |
0.1205950675 |
- |
- |
hypothetical protein JCGZ_06749 [Jatropha curcas] |
| 14 |
Hb_000753_200 |
0.1207184359 |
- |
- |
PREDICTED: putative ATP-dependent helicase hrq1 [Jatropha curcas] |
| 15 |
Hb_001489_050 |
0.1238360125 |
- |
- |
PREDICTED: protein TWIN LOV 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000540_150 |
0.1241871187 |
- |
- |
PREDICTED: uncharacterized protein YKR070W [Jatropha curcas] |
| 17 |
Hb_000740_050 |
0.1245087416 |
- |
- |
PREDICTED: probable lysine--tRNA ligase, cytoplasmic isoform X2 [Jatropha curcas] |
| 18 |
Hb_035755_010 |
0.1258438313 |
- |
- |
unknown [Populus trichocarpa] |
| 19 |
Hb_000367_130 |
0.1266470389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_153533_010 |
0.1270479129 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X3 [Jatropha curcas] |