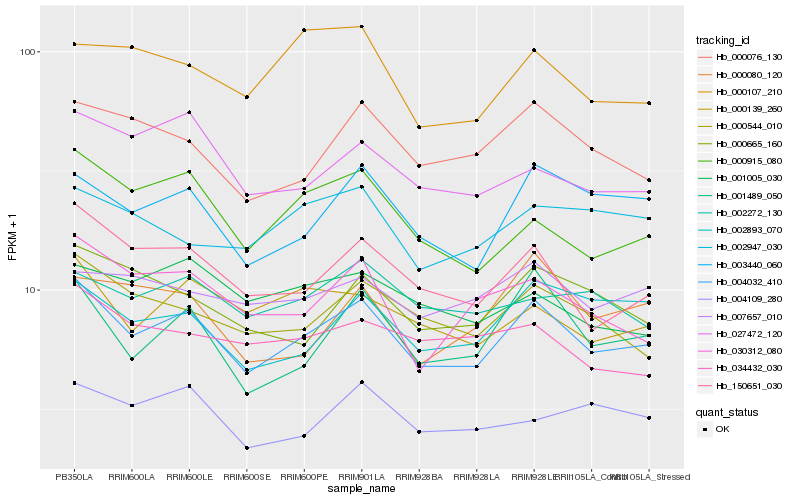
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004032_410 |
0.0 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 2 |
Hb_150651_030 |
0.0725938312 |
- |
- |
PREDICTED: metallophosphoesterase 1-like [Jatropha curcas] |
| 3 |
Hb_003440_060 |
0.0802969561 |
- |
- |
PREDICTED: translin-associated protein X isoform X1 [Jatropha curcas] |
| 4 |
Hb_001489_050 |
0.0809918121 |
- |
- |
PREDICTED: protein TWIN LOV 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000544_010 |
0.0828070214 |
- |
- |
PREDICTED: GPI mannosyltransferase 1 [Jatropha curcas] |
| 6 |
Hb_000076_130 |
0.0850016165 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 7 |
Hb_000915_080 |
0.0867831526 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 8 |
Hb_000665_160 |
0.0906468704 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000139_260 |
0.0910053309 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 10 |
Hb_002893_070 |
0.0911549928 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_030312_080 |
0.0917629354 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 12 |
Hb_027472_120 |
0.0920072822 |
- |
- |
PREDICTED: thiosulfate/3-mercaptopyruvate sulfurtransferase 1, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_004109_280 |
0.0967765086 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 14 |
Hb_002272_130 |
0.0975476852 |
- |
- |
PREDICTED: DNA excision repair protein ERCC-1 [Jatropha curcas] |
| 15 |
Hb_007657_010 |
0.0986136204 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_000080_120 |
0.0998568059 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350-like [Jatropha curcas] |
| 17 |
Hb_034432_030 |
0.1000553426 |
- |
- |
PREDICTED: probable glycosyltransferase At5g20260 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000107_210 |
0.1011349731 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 19 |
Hb_001005_030 |
0.1013243094 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 20 |
Hb_002947_030 |
0.1014943135 |
- |
- |
PREDICTED: uncharacterized protein LOC105633529 [Jatropha curcas] |