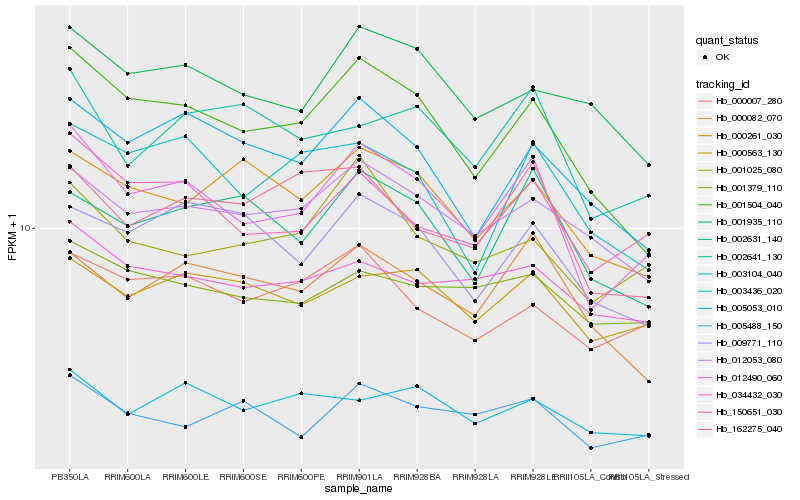
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012490_060 |
0.0 |
- |
- |
PREDICTED: UBP1-associated proteins 1C isoform X1 [Jatropha curcas] |
| 2 |
Hb_150651_030 |
0.0800592385 |
- |
- |
PREDICTED: metallophosphoesterase 1-like [Jatropha curcas] |
| 3 |
Hb_001504_040 |
0.0825023651 |
- |
- |
PREDICTED: factor of DNA methylation 1-like [Jatropha curcas] |
| 4 |
Hb_005488_150 |
0.10334273 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_000563_130 |
0.1035145277 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 6 |
Hb_012053_080 |
0.105493211 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 7 |
Hb_002641_130 |
0.1057001948 |
- |
- |
PREDICTED: uncharacterized protein LOC105643556 [Jatropha curcas] |
| 8 |
Hb_005053_010 |
0.1059328038 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 9 |
Hb_001025_080 |
0.1072310169 |
- |
- |
hypothetical protein JCGZ_11431 [Jatropha curcas] |
| 10 |
Hb_002631_140 |
0.1087730451 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 12, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000007_280 |
0.1091605461 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 12 |
Hb_034432_030 |
0.109402914 |
- |
- |
PREDICTED: probable glycosyltransferase At5g20260 isoform X1 [Jatropha curcas] |
| 13 |
Hb_162275_040 |
0.1094405916 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003104_040 |
0.1096922676 |
- |
- |
PREDICTED: insulin-degrading enzyme isoform X1 [Jatropha curcas] |
| 15 |
Hb_000082_070 |
0.1103338001 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 1 [Jatropha curcas] |
| 16 |
Hb_001379_110 |
0.1111217308 |
- |
- |
PREDICTED: sec1 family domain-containing protein MIP3 [Jatropha curcas] |
| 17 |
Hb_003436_020 |
0.1116668197 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000261_030 |
0.1120156648 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 19 |
Hb_009771_110 |
0.1120887939 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 2 [Jatropha curcas] |
| 20 |
Hb_001935_110 |
0.1135874871 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |