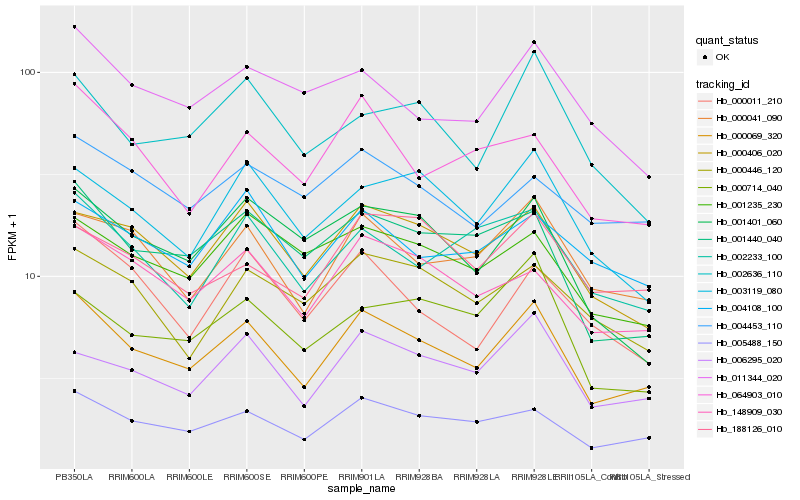
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_320 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642846 [Jatropha curcas] |
| 2 |
Hb_005488_150 |
0.0771849514 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000041_090 |
0.0887211951 |
- |
- |
PREDICTED: uncharacterized protein LOC105641452 [Jatropha curcas] |
| 4 |
Hb_148909_030 |
0.0908038302 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000406_020 |
0.0935597808 |
- |
- |
- |
| 6 |
Hb_006295_020 |
0.0945974631 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATR isoform X1 [Jatropha curcas] |
| 7 |
Hb_001440_040 |
0.0953804553 |
- |
- |
hypothetical protein POPTR_0008s03520g [Populus trichocarpa] |
| 8 |
Hb_003119_080 |
0.1044506312 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 9 |
Hb_002233_100 |
0.1058140708 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 6-like [Malus domestica] |
| 10 |
Hb_064903_010 |
0.1075421239 |
- |
- |
PREDICTED: T-complex protein 1 subunit delta [Jatropha curcas] |
| 11 |
Hb_001235_230 |
0.1078569164 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 12 |
Hb_001401_060 |
0.1081266641 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 13 |
Hb_004108_100 |
0.1088666471 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X2 [Jatropha curcas] |
| 14 |
Hb_002636_110 |
0.1090830275 |
- |
- |
PREDICTED: heat shock protein 83-like isoform X2 [Gossypium raimondii] |
| 15 |
Hb_011344_020 |
0.1099132656 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 16 |
Hb_000011_210 |
0.112413252 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 17 |
Hb_004453_110 |
0.1128502626 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 18 |
Hb_000714_040 |
0.1129660826 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000446_120 |
0.1134912049 |
- |
- |
PREDICTED: uncharacterized transmembrane protein DDB_G0289901 [Jatropha curcas] |
| 20 |
Hb_188126_010 |
0.113538846 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like isoform X3 [Jatropha curcas] |