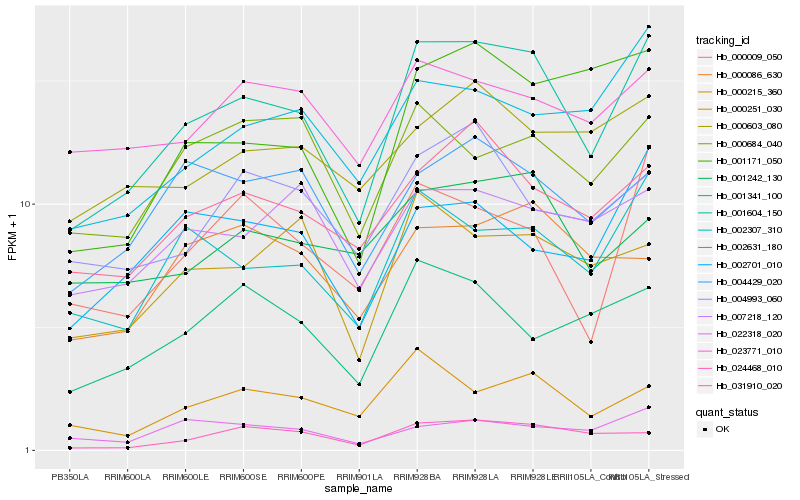
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001604_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial-like [Nelumbo nucifera] |
| 2 |
Hb_002307_310 |
0.1139144688 |
- |
- |
PREDICTED: outer envelope pore protein 37, chloroplastic [Jatropha curcas] |
| 3 |
Hb_031910_020 |
0.1213379211 |
- |
- |
PREDICTED: exocyst complex component SEC15A [Jatropha curcas] |
| 4 |
Hb_002631_180 |
0.1353493442 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-2 [Jatropha curcas] |
| 5 |
Hb_000215_360 |
0.1355638853 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 6 |
Hb_000009_050 |
0.136613749 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 7 |
Hb_004429_020 |
0.1375685907 |
- |
- |
PREDICTED: magnesium transporter MRS2-5 [Jatropha curcas] |
| 8 |
Hb_000251_030 |
0.1382091825 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_001242_130 |
0.1385454252 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 4-like [Jatropha curcas] |
| 10 |
Hb_002701_010 |
0.1401085538 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001171_050 |
0.1407989092 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0284459 [Jatropha curcas] |
| 12 |
Hb_007218_120 |
0.1417224267 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 13 |
Hb_001341_100 |
0.1433833259 |
- |
- |
hypothetical protein JCGZ_05568 [Jatropha curcas] |
| 14 |
Hb_004993_060 |
0.1434927787 |
- |
- |
- |
| 15 |
Hb_000603_080 |
0.1458916134 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33170 [Vitis vinifera] |
| 16 |
Hb_024468_010 |
0.1461248292 |
- |
- |
hypothetical protein CISIN_1g046520mg [Citrus sinensis] |
| 17 |
Hb_023771_010 |
0.1462041842 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000684_040 |
0.1466579354 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 19 |
Hb_022318_020 |
0.1474636569 |
- |
- |
hypothetical protein JCGZ_00541 [Jatropha curcas] |
| 20 |
Hb_000086_630 |
0.1480339294 |
- |
- |
PREDICTED: dicarboxylate transporter 2.1, chloroplastic [Jatropha curcas] |