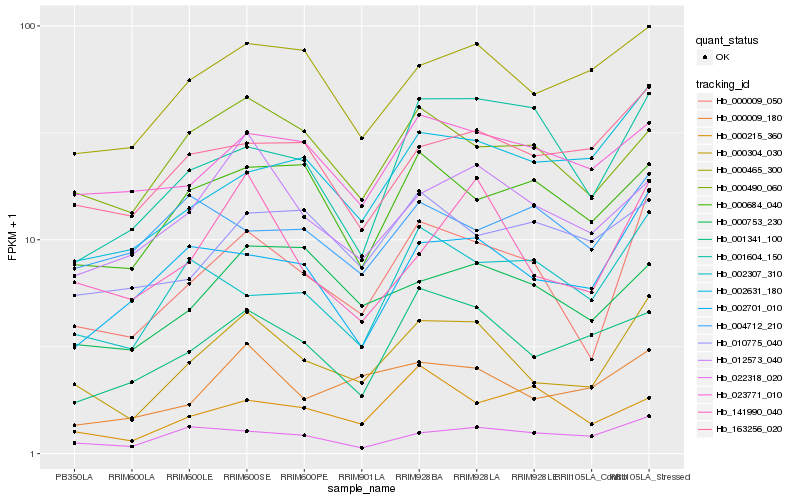
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000009_050 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 2 |
Hb_000304_030 |
0.1036876459 |
- |
- |
PREDICTED: seed maturation protein PM36 [Jatropha curcas] |
| 3 |
Hb_001604_150 |
0.136613749 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial-like [Nelumbo nucifera] |
| 4 |
Hb_002701_010 |
0.1389243977 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002307_310 |
0.1435004065 |
- |
- |
PREDICTED: outer envelope pore protein 37, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000684_040 |
0.1554582073 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 7 |
Hb_002631_180 |
0.1559291916 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-2 [Jatropha curcas] |
| 8 |
Hb_000490_060 |
0.1562049365 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 9 |
Hb_023771_010 |
0.157041209 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 10 |
Hb_163256_020 |
0.1624753504 |
- |
- |
fructokinase [Manihot esculenta] |
| 11 |
Hb_022318_020 |
0.1643204758 |
- |
- |
hypothetical protein JCGZ_00541 [Jatropha curcas] |
| 12 |
Hb_010775_040 |
0.1649582377 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 13 |
Hb_000215_360 |
0.167720731 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 14 |
Hb_012573_040 |
0.1682681606 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g55270-like [Jatropha curcas] |
| 15 |
Hb_141990_040 |
0.1698926517 |
- |
- |
Protein SCAR2, putative [Ricinus communis] |
| 16 |
Hb_000465_300 |
0.169999563 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 17 |
Hb_000753_230 |
0.1710185328 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000009_180 |
0.1714023997 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
RecName: Full=Geranylgeranyl pyrophosphate synthase, chloroplastic; Short=GGPP synthase; Short=GGPS; AltName: Full=(2E,6E)-farnesyl diphosphate synthase; AltName: Full=Dimethylallyltranstransferase; AltName: Full=Farnesyl diphosphate synthase; AltName: Full=Farnesyltranstransferase; AltName: Full=Geranyltranstransferase; Flags: Precursor [Hevea brasiliensis] |
| 19 |
Hb_004712_210 |
0.1715475368 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM50-like [Jatropha curcas] |
| 20 |
Hb_001341_100 |
0.1721854318 |
- |
- |
hypothetical protein JCGZ_05568 [Jatropha curcas] |