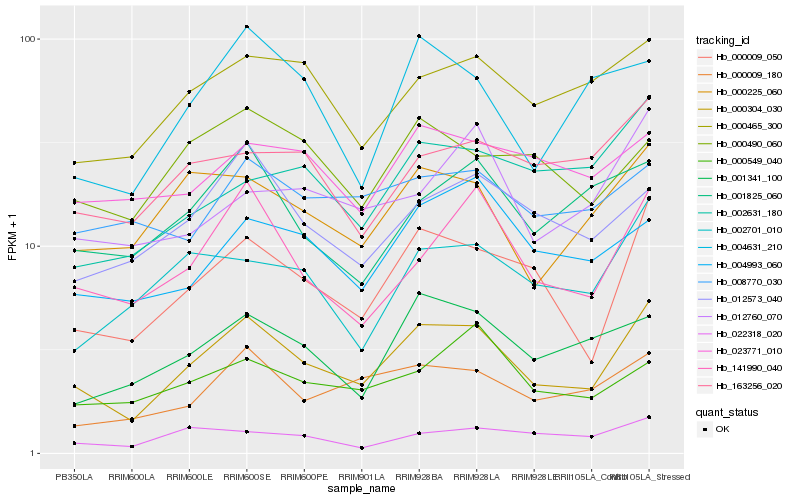
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000304_030 |
0.0 |
- |
- |
PREDICTED: seed maturation protein PM36 [Jatropha curcas] |
| 2 |
Hb_000009_050 |
0.1036876459 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 3 |
Hb_141990_040 |
0.1312184402 |
- |
- |
Protein SCAR2, putative [Ricinus communis] |
| 4 |
Hb_000009_180 |
0.1353309605 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
RecName: Full=Geranylgeranyl pyrophosphate synthase, chloroplastic; Short=GGPP synthase; Short=GGPS; AltName: Full=(2E,6E)-farnesyl diphosphate synthase; AltName: Full=Dimethylallyltranstransferase; AltName: Full=Farnesyl diphosphate synthase; AltName: Full=Farnesyltranstransferase; AltName: Full=Geranyltranstransferase; Flags: Precursor [Hevea brasiliensis] |
| 5 |
Hb_000225_060 |
0.1364137002 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 6 |
Hb_000465_300 |
0.1434483189 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 7 |
Hb_001341_100 |
0.146090308 |
- |
- |
hypothetical protein JCGZ_05568 [Jatropha curcas] |
| 8 |
Hb_012573_040 |
0.1479200063 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g55270-like [Jatropha curcas] |
| 9 |
Hb_004993_060 |
0.1507786259 |
- |
- |
- |
| 10 |
Hb_002701_010 |
0.1515913294 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004631_210 |
0.1517682959 |
- |
- |
hypothetical protein POPTR_0007s05070g [Populus trichocarpa] |
| 12 |
Hb_001825_060 |
0.1524753553 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 13 |
Hb_163256_020 |
0.1558302572 |
- |
- |
fructokinase [Manihot esculenta] |
| 14 |
Hb_000490_060 |
0.1573064478 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 15 |
Hb_000549_040 |
0.1581332191 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_022318_020 |
0.1599718199 |
- |
- |
hypothetical protein JCGZ_00541 [Jatropha curcas] |
| 17 |
Hb_012760_070 |
0.160747675 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 18 |
Hb_023771_010 |
0.1621169024 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 19 |
Hb_002631_180 |
0.1624402951 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-2 [Jatropha curcas] |
| 20 |
Hb_008770_030 |
0.1625653816 |
- |
- |
PREDICTED: MACPF domain-containing protein At4g24290-like [Jatropha curcas] |