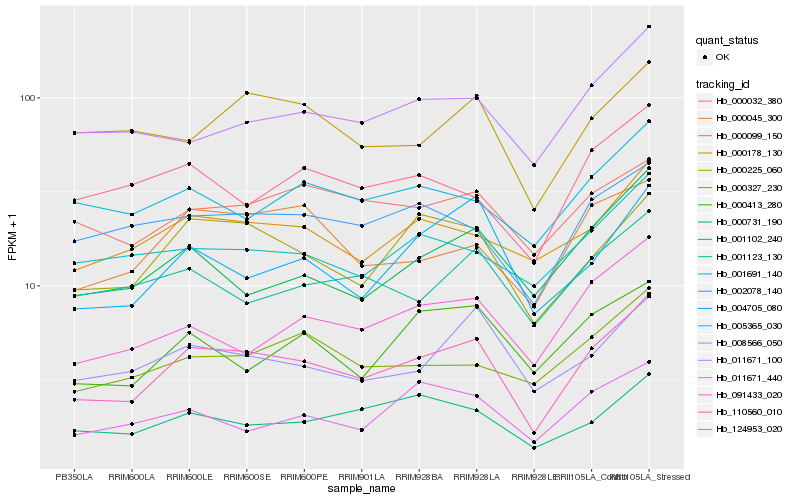
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_110560_010 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_000225_060 |
0.1025348286 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 3 |
Hb_008566_050 |
0.117424954 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 4 |
Hb_004705_080 |
0.1180557257 |
transcription factor |
TF Family: ARF |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000099_150 |
0.1196152245 |
- |
- |
PREDICTED: calcium-dependent protein kinase 3 [Jatropha curcas] |
| 6 |
Hb_000045_300 |
0.1202062086 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier isoform X1 [Jatropha curcas] |
| 7 |
Hb_091433_020 |
0.1208340348 |
- |
- |
ribosomal RNA methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_005365_030 |
0.1214078301 |
- |
- |
hypothetical protein POPTR_0002s24010g [Populus trichocarpa] |
| 9 |
Hb_001102_240 |
0.1235480657 |
- |
- |
PREDICTED: NADH kinase isoform X1 [Jatropha curcas] |
| 10 |
Hb_011671_100 |
0.1247772648 |
- |
- |
60S ribosomal protein L15 [Populus trichocarpa] |
| 11 |
Hb_000731_190 |
0.1260124689 |
- |
- |
PREDICTED: cardiolipin synthase (CMP-forming), mitochondrial [Jatropha curcas] |
| 12 |
Hb_000327_230 |
0.1279310879 |
- |
- |
PREDICTED: nitrilase-like protein 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000413_280 |
0.1292611109 |
- |
- |
PREDICTED: probable UDP-3-O-acylglucosamine N-acyltransferase 2, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000178_130 |
0.1294834292 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 15 |
Hb_001691_140 |
0.1301988503 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |
| 16 |
Hb_011671_440 |
0.1304883613 |
- |
- |
ATP-dependent DNA helicase pcrA, putative [Ricinus communis] |
| 17 |
Hb_001123_130 |
0.1311571751 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_000032_380 |
0.1312136921 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002078_140 |
0.1321810705 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 20 |
Hb_124953_020 |
0.1321929981 |
- |
- |
Mitochondrial import inner membrane translocase subunit Tim13, putative [Ricinus communis] |