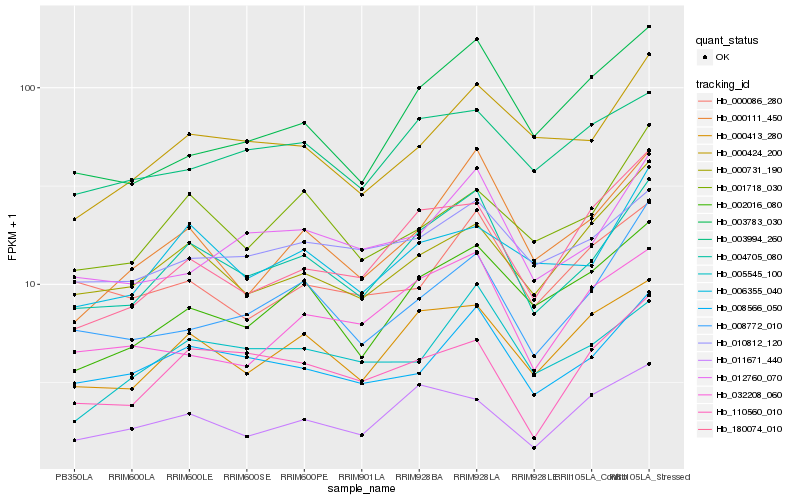
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004705_080 |
0.0 |
transcription factor |
TF Family: ARF |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_000413_280 |
0.0903879586 |
- |
- |
PREDICTED: probable UDP-3-O-acylglucosamine N-acyltransferase 2, mitochondrial [Jatropha curcas] |
| 3 |
Hb_008566_050 |
0.0937065622 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 4 |
Hb_000111_450 |
0.0972530948 |
- |
- |
PREDICTED: probable choline kinase 2 [Jatropha curcas] |
| 5 |
Hb_012760_070 |
0.0999703026 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 6 |
Hb_002016_080 |
0.1061152498 |
- |
- |
PREDICTED: thiamine pyrophosphokinase 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000424_200 |
0.1082583285 |
- |
- |
hypothetical protein JCGZ_07583 [Jatropha curcas] |
| 8 |
Hb_006355_040 |
0.1098689566 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_010812_120 |
0.112066296 |
- |
- |
hypothetical protein JCGZ_03293 [Jatropha curcas] |
| 10 |
Hb_005545_100 |
0.112371883 |
- |
- |
PREDICTED: uncharacterized protein LOC105645673 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003783_030 |
0.1157837974 |
- |
- |
hypothetical protein POPTR_0018s13980g [Populus trichocarpa] |
| 12 |
Hb_008772_010 |
0.1161742485 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_110560_010 |
0.1180557257 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_003994_260 |
0.1195067137 |
- |
- |
PREDICTED: uncharacterized protein LOC105646151 [Jatropha curcas] |
| 15 |
Hb_011671_440 |
0.1196712929 |
- |
- |
ATP-dependent DNA helicase pcrA, putative [Ricinus communis] |
| 16 |
Hb_000731_190 |
0.120451175 |
- |
- |
PREDICTED: cardiolipin synthase (CMP-forming), mitochondrial [Jatropha curcas] |
| 17 |
Hb_001718_030 |
0.1208338195 |
- |
- |
PREDICTED: calcineurin B-like protein 9 isoform X1 [Jatropha curcas] |
| 18 |
Hb_032208_060 |
0.1226559346 |
- |
- |
PREDICTED: constitutive photomorphogenesis protein 10 isoform X2 [Populus euphratica] |
| 19 |
Hb_000086_280 |
0.123267506 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 2 homolog 3-like [Gossypium raimondii] |
| 20 |
Hb_180074_010 |
0.1237351581 |
- |
- |
Single-stranded DNA-binding protein, mitochondrial precursor, putative [Ricinus communis] |