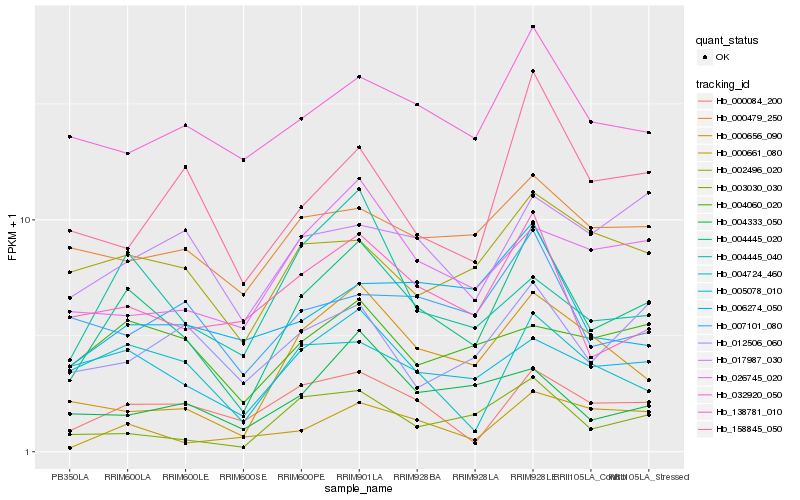
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004445_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_13166 [Jatropha curcas] |
| 2 |
Hb_005078_010 |
0.1562219599 |
- |
- |
Derlin-2, putative [Ricinus communis] |
| 3 |
Hb_000084_200 |
0.1611475438 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X2 [Jatropha curcas] |
| 4 |
Hb_138781_010 |
0.1649655506 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 5 |
Hb_003030_030 |
0.1653952772 |
- |
- |
- |
| 6 |
Hb_032920_050 |
0.1771316371 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_026745_020 |
0.1799194912 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 8, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000661_080 |
0.1817874815 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 9 |
Hb_002496_020 |
0.1825076977 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_004724_460 |
0.1831386976 |
- |
- |
hypothetical protein EUGRSUZ_L02100 [Eucalyptus grandis] |
| 11 |
Hb_004060_020 |
0.1838295923 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_004445_040 |
0.1861127231 |
- |
- |
PREDICTED: uncharacterized protein LOC105639134 isoform X5 [Jatropha curcas] |
| 13 |
Hb_012506_060 |
0.1872502659 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |
| 14 |
Hb_158845_050 |
0.1874970043 |
- |
- |
PREDICTED: uncharacterized protein LOC105122890 [Populus euphratica] |
| 15 |
Hb_004333_050 |
0.1884701016 |
- |
- |
- |
| 16 |
Hb_000479_250 |
0.1887096292 |
- |
- |
PREDICTED: uncharacterized protein LOC105644121 [Jatropha curcas] |
| 17 |
Hb_007101_080 |
0.1893245624 |
transcription factor |
TF Family: GNAT |
GCN5-related N-acetyltransferase family protein [Populus trichocarpa] |
| 18 |
Hb_017987_030 |
0.1902724309 |
- |
- |
PREDICTED: uncharacterized protein LOC105630796 [Jatropha curcas] |
| 19 |
Hb_006274_050 |
0.1913859002 |
- |
- |
hypothetical protein VITISV_022618 [Vitis vinifera] |
| 20 |
Hb_000656_090 |
0.1922913609 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |