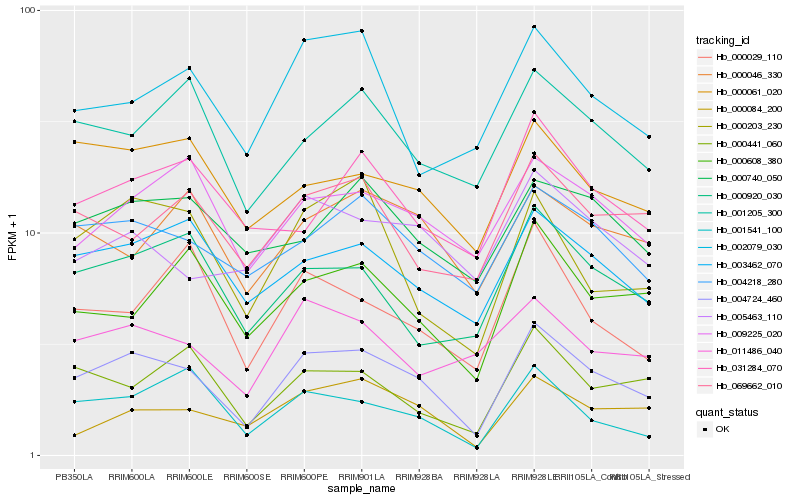
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004724_460 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_L02100 [Eucalyptus grandis] |
| 2 |
Hb_004218_280 |
0.1304190277 |
- |
- |
Exoenzymes regulatory protein aepA precursor, putative [Ricinus communis] |
| 3 |
Hb_003462_070 |
0.1341083641 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 4 |
Hb_000203_230 |
0.1397068244 |
- |
- |
PREDICTED: probable inactive dual specificity protein phosphatase-like At4g18593 [Jatropha curcas] |
| 5 |
Hb_000084_200 |
0.1420767331 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000920_030 |
0.1436829886 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_000061_020 |
0.1462570836 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02 [Jatropha curcas] |
| 8 |
Hb_000608_380 |
0.1463212396 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001205_300 |
0.1464536847 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 10 |
Hb_009225_020 |
0.1468890098 |
- |
- |
PREDICTED: uncharacterized protein LOC105638232 [Jatropha curcas] |
| 11 |
Hb_000441_060 |
0.1492636582 |
- |
- |
PREDICTED: glycolipid transfer protein [Jatropha curcas] |
| 12 |
Hb_001541_100 |
0.1506277774 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |
| 13 |
Hb_069662_010 |
0.1507234726 |
- |
- |
PREDICTED: uncharacterized protein LOC103320051 [Prunus mume] |
| 14 |
Hb_011486_040 |
0.1509294774 |
- |
- |
PREDICTED: protein trichome birefringence-like 13 [Jatropha curcas] |
| 15 |
Hb_000046_330 |
0.1516825009 |
- |
- |
PREDICTED: uncharacterized protein LOC105631826 [Jatropha curcas] |
| 16 |
Hb_005463_110 |
0.1517927773 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase HPR3-like [Jatropha curcas] |
| 17 |
Hb_000740_050 |
0.151894806 |
- |
- |
PREDICTED: probable lysine--tRNA ligase, cytoplasmic isoform X2 [Jatropha curcas] |
| 18 |
Hb_002079_030 |
0.1523322123 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000029_110 |
0.1552883875 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_031284_070 |
0.1553146884 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |