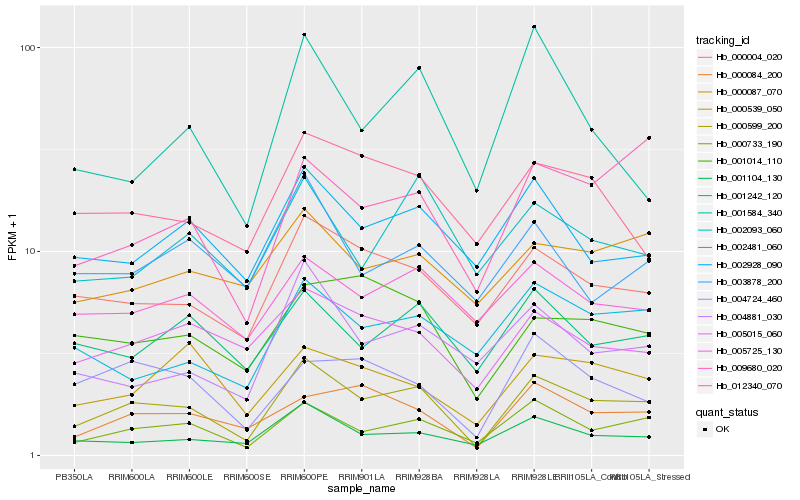
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000599_200 |
0.0 |
- |
- |
ABC transporter family protein [Populus trichocarpa] |
| 2 |
Hb_000733_190 |
0.1121101165 |
- |
- |
zinc ion binding protein, putative [Ricinus communis] |
| 3 |
Hb_001104_130 |
0.1422324983 |
- |
- |
C-14 sterol reductase, putative [Ricinus communis] |
| 4 |
Hb_000004_020 |
0.1452620733 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000084_200 |
0.1513342544 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X2 [Jatropha curcas] |
| 6 |
Hb_009680_020 |
0.1616268431 |
- |
- |
PREDICTED: 40S ribosomal protein S10 [Jatropha curcas] |
| 7 |
Hb_002928_090 |
0.1618852551 |
- |
- |
PREDICTED: metallocarboxypeptidase A-like protein TRV_02598 [Jatropha curcas] |
| 8 |
Hb_001242_120 |
0.1627803332 |
- |
- |
PREDICTED: carboxypeptidase D [Jatropha curcas] |
| 9 |
Hb_005015_060 |
0.1632857327 |
- |
- |
PREDICTED: developmentally-regulated G-protein 3 [Jatropha curcas] |
| 10 |
Hb_001014_110 |
0.1641825193 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 11 |
Hb_004724_460 |
0.1645813152 |
- |
- |
hypothetical protein EUGRSUZ_L02100 [Eucalyptus grandis] |
| 12 |
Hb_004881_030 |
0.1665972817 |
- |
- |
PREDICTED: uncharacterized protein LOC105629532 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000539_050 |
0.1685850533 |
- |
- |
bile acid:sodium symporter, putative [Ricinus communis] |
| 14 |
Hb_005725_130 |
0.1691866437 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 15 |
Hb_002481_060 |
0.1722233525 |
- |
- |
PREDICTED: uncharacterized protein LOC105640851 [Jatropha curcas] |
| 16 |
Hb_002093_060 |
0.17244271 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 17 |
Hb_012340_070 |
0.172699811 |
- |
- |
Gamma-secretase subunit APH1-like family protein [Populus trichocarpa] |
| 18 |
Hb_000087_070 |
0.1747025406 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 19 |
Hb_003878_200 |
0.1760588579 |
- |
- |
PREDICTED: V-type proton ATPase subunit D-like [Jatropha curcas] |
| 20 |
Hb_001584_340 |
0.1761487398 |
- |
- |
PREDICTED: ATP-citrate synthase alpha chain protein 2 [Jatropha curcas] |