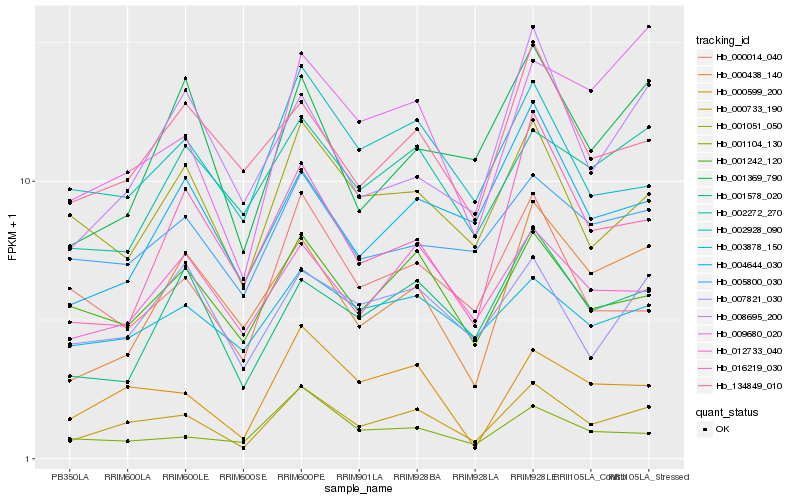
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000733_190 |
0.0 |
- |
- |
zinc ion binding protein, putative [Ricinus communis] |
| 2 |
Hb_000599_200 |
0.1121101165 |
- |
- |
ABC transporter family protein [Populus trichocarpa] |
| 3 |
Hb_001242_120 |
0.1239992345 |
- |
- |
PREDICTED: carboxypeptidase D [Jatropha curcas] |
| 4 |
Hb_001051_050 |
0.1262276351 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_009680_020 |
0.1290514121 |
- |
- |
PREDICTED: 40S ribosomal protein S10 [Jatropha curcas] |
| 6 |
Hb_012733_040 |
0.1304081792 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 7 |
Hb_002928_090 |
0.1336240844 |
- |
- |
PREDICTED: metallocarboxypeptidase A-like protein TRV_02598 [Jatropha curcas] |
| 8 |
Hb_000438_140 |
0.1351093739 |
- |
- |
hypothetical protein CISIN_1g018088mg [Citrus sinensis] |
| 9 |
Hb_007821_030 |
0.1352563599 |
- |
- |
5'-nucleotidase domain-containing [Gossypium arboreum] |
| 10 |
Hb_134849_010 |
0.1364645589 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 11 |
Hb_001578_020 |
0.1364751779 |
- |
- |
Protein virR, putative [Ricinus communis] |
| 12 |
Hb_001369_790 |
0.1375214387 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic [Jatropha curcas] |
| 13 |
Hb_005800_030 |
0.1381653141 |
- |
- |
PREDICTED: lysM and putative peptidoglycan-binding domain-containing protein 4-like [Jatropha curcas] |
| 14 |
Hb_008695_200 |
0.1404190741 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 15 |
Hb_004644_030 |
0.1417083563 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 16 |
Hb_003878_150 |
0.1417490612 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 17 |
Hb_000014_040 |
0.142338071 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002272_270 |
0.1424202655 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 19 |
Hb_016219_030 |
0.1427632668 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_001104_130 |
0.1440967796 |
- |
- |
C-14 sterol reductase, putative [Ricinus communis] |