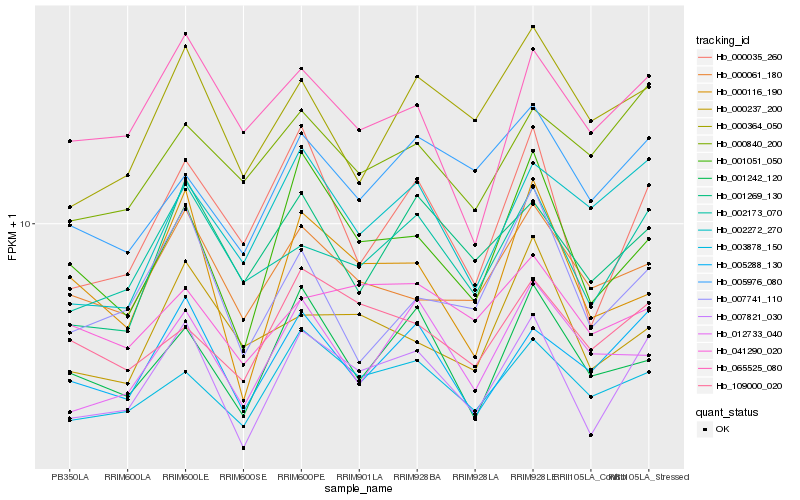
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007821_030 |
0.0 |
- |
- |
5'-nucleotidase domain-containing [Gossypium arboreum] |
| 2 |
Hb_002173_070 |
0.0810104276 |
- |
- |
MRG family protein isoform 2 [Theobroma cacao] |
| 3 |
Hb_005288_130 |
0.0874591238 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_001051_050 |
0.0900422054 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003878_150 |
0.0946114611 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 6 |
Hb_000035_260 |
0.0955613829 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 7 |
Hb_000061_180 |
0.1033277767 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 8 |
Hb_001269_130 |
0.1043929217 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 9 |
Hb_001242_120 |
0.1051898573 |
- |
- |
PREDICTED: carboxypeptidase D [Jatropha curcas] |
| 10 |
Hb_109000_020 |
0.1063743195 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 11 |
Hb_005976_080 |
0.1068584842 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 12 |
Hb_000116_190 |
0.1072830274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_012733_040 |
0.1073728979 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 14 |
Hb_000364_050 |
0.1102957078 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_041290_020 |
0.1111228986 |
- |
- |
PREDICTED: centromere/kinetochore protein zw10 homolog isoform X2 [Jatropha curcas] |
| 16 |
Hb_065525_080 |
0.1111235919 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 14 [Populus euphratica] |
| 17 |
Hb_002272_270 |
0.1112174301 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 18 |
Hb_000840_200 |
0.1117572899 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase small chain, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_000237_200 |
0.112922422 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 20 |
Hb_007741_110 |
0.1135389304 |
- |
- |
- |