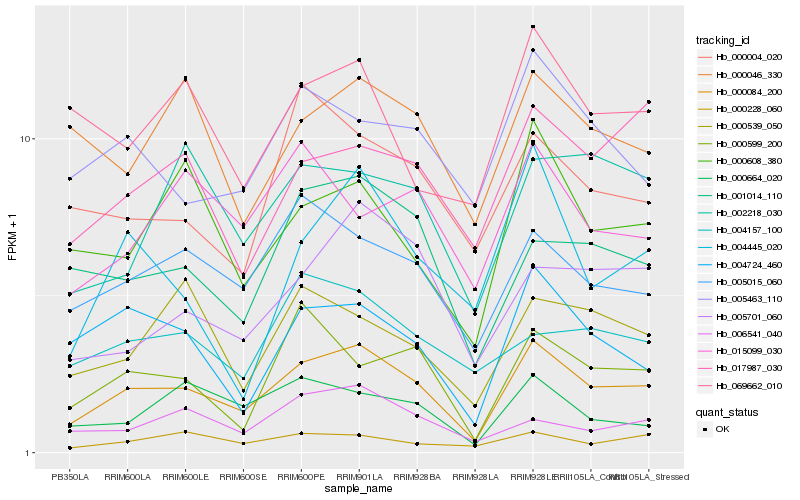
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000084_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X2 [Jatropha curcas] |
| 2 |
Hb_005015_060 |
0.1364178928 |
- |
- |
PREDICTED: developmentally-regulated G-protein 3 [Jatropha curcas] |
| 3 |
Hb_001014_110 |
0.1405007628 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 4 |
Hb_004724_460 |
0.1420767331 |
- |
- |
hypothetical protein EUGRSUZ_L02100 [Eucalyptus grandis] |
| 5 |
Hb_000608_380 |
0.1441952558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005701_060 |
0.1493135714 |
- |
- |
hypothetical protein JCGZ_04513 [Jatropha curcas] |
| 7 |
Hb_000599_200 |
0.1513342544 |
- |
- |
ABC transporter family protein [Populus trichocarpa] |
| 8 |
Hb_000539_050 |
0.1543686669 |
- |
- |
bile acid:sodium symporter, putative [Ricinus communis] |
| 9 |
Hb_005463_110 |
0.1556913826 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase HPR3-like [Jatropha curcas] |
| 10 |
Hb_017987_030 |
0.1561110415 |
- |
- |
PREDICTED: uncharacterized protein LOC105630796 [Jatropha curcas] |
| 11 |
Hb_002218_030 |
0.1564577716 |
- |
- |
hypothetical protein B456_007G345200 [Gossypium raimondii] |
| 12 |
Hb_000664_020 |
0.1565588899 |
- |
- |
tryptophanyl-tRNA synthetase, putative [Ricinus communis] |
| 13 |
Hb_000228_060 |
0.1565649909 |
- |
- |
hypothetical protein JCGZ_21532 [Jatropha curcas] |
| 14 |
Hb_006541_040 |
0.1584550872 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 2 [Jatropha curcas] |
| 15 |
Hb_000004_020 |
0.1594285259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004445_020 |
0.1611475438 |
- |
- |
hypothetical protein JCGZ_13166 [Jatropha curcas] |
| 17 |
Hb_004157_100 |
0.1625360234 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 18 |
Hb_015099_030 |
0.162764814 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 19 |
Hb_069662_010 |
0.1638465789 |
- |
- |
PREDICTED: uncharacterized protein LOC103320051 [Prunus mume] |
| 20 |
Hb_000046_330 |
0.1650049853 |
- |
- |
PREDICTED: uncharacterized protein LOC105631826 [Jatropha curcas] |