| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
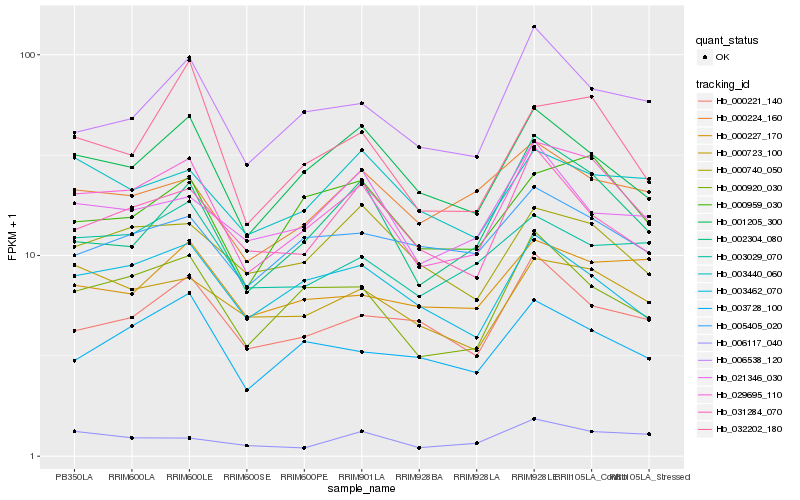
Hb_029695_110 |
0.0 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 2 |
Hb_000920_030 |
0.086261163 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_001205_300 |
0.0915571399 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 4 |
Hb_000959_030 |
0.0996742229 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 5 |
Hb_003462_070 |
0.1014249707 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 6 |
Hb_002304_080 |
0.1018170651 |
- |
- |
PREDICTED: uncharacterized protein LOC105649626 [Jatropha curcas] |
| 7 |
Hb_000723_100 |
0.1044972366 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 8 |
Hb_032202_180 |
0.104958918 |
- |
- |
PREDICTED: protein-ribulosamine 3-kinase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_006538_120 |
0.1058997675 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 10 |
Hb_031284_070 |
0.1095179786 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000740_050 |
0.1121448042 |
- |
- |
PREDICTED: probable lysine--tRNA ligase, cytoplasmic isoform X2 [Jatropha curcas] |
| 12 |
Hb_003728_100 |
0.1145503654 |
transcription factor |
TF Family: FAR1 |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000221_140 |
0.1148704404 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000224_160 |
0.1174417381 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 15 |
Hb_003029_070 |
0.1186548441 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_005405_020 |
0.1203980293 |
- |
- |
PREDICTED: petal death protein isoform X1 [Jatropha curcas] |
| 17 |
Hb_021346_030 |
0.1221337679 |
- |
- |
PREDICTED: uncharacterized protein LOC105141875 [Populus euphratica] |
| 18 |
Hb_003440_060 |
0.1224561826 |
- |
- |
PREDICTED: translin-associated protein X isoform X1 [Jatropha curcas] |
| 19 |
Hb_006117_040 |
0.1229291595 |
- |
- |
PREDICTED: seed biotin-containing protein SBP65 [Jatropha curcas] |
| 20 |
Hb_000227_170 |
0.123703143 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |