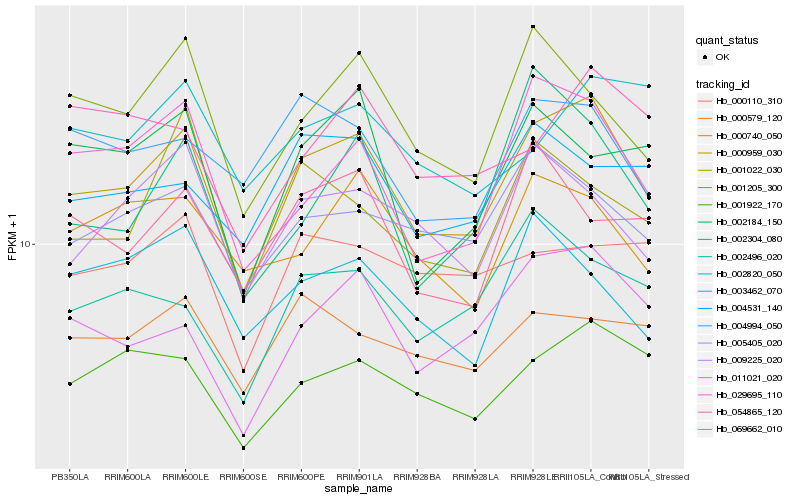
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000959_030 |
0.0 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 2 |
Hb_011021_020 |
0.0961463634 |
- |
- |
PREDICTED: uncharacterized protein LOC105644392 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001922_170 |
0.0995898298 |
- |
- |
Plastidic pyruvate kinase beta subunit 1 isoform 2 [Theobroma cacao] |
| 4 |
Hb_029695_110 |
0.0996742229 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 5 |
Hb_005405_020 |
0.1060326629 |
- |
- |
PREDICTED: petal death protein isoform X1 [Jatropha curcas] |
| 6 |
Hb_001205_300 |
0.1087598648 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 7 |
Hb_009225_020 |
0.1106596459 |
- |
- |
PREDICTED: uncharacterized protein LOC105638232 [Jatropha curcas] |
| 8 |
Hb_004994_050 |
0.1133606487 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 9 |
Hb_000740_050 |
0.1145951191 |
- |
- |
PREDICTED: probable lysine--tRNA ligase, cytoplasmic isoform X2 [Jatropha curcas] |
| 10 |
Hb_000579_120 |
0.1161342728 |
- |
- |
PREDICTED: uncharacterized protein LOC105633845 [Jatropha curcas] |
| 11 |
Hb_002496_020 |
0.1170924122 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_004531_140 |
0.1179810781 |
- |
- |
PREDICTED: uncharacterized protein LOC105635023 [Jatropha curcas] |
| 13 |
Hb_069662_010 |
0.1216519714 |
- |
- |
PREDICTED: uncharacterized protein LOC103320051 [Prunus mume] |
| 14 |
Hb_000110_310 |
0.1221108236 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 4 of pyruvate dehydrogenase complex |
Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_002184_150 |
0.1230735479 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_001022_030 |
0.1232011403 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 17 |
Hb_002304_080 |
0.1237410805 |
- |
- |
PREDICTED: uncharacterized protein LOC105649626 [Jatropha curcas] |
| 18 |
Hb_003462_070 |
0.1239851189 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 19 |
Hb_054865_120 |
0.1240112035 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 20 |
Hb_002820_050 |
0.1257742847 |
- |
- |
PREDICTED: putative glutathione-specific gamma-glutamylcyclotransferase 2 [Jatropha curcas] |