| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
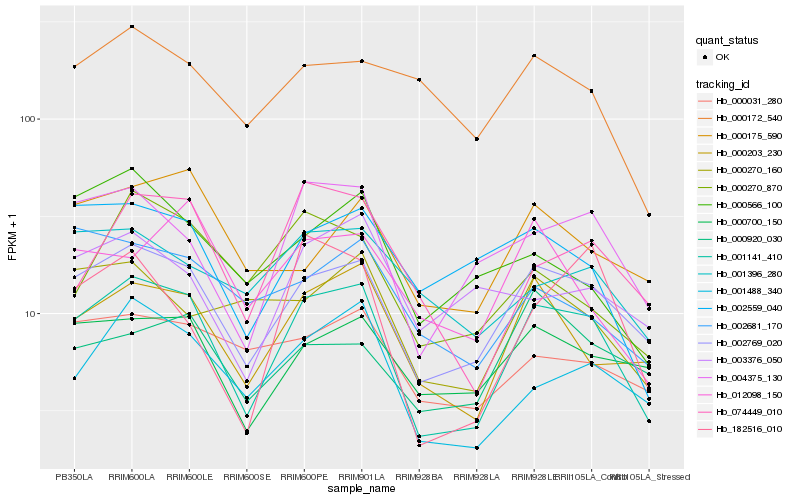
Hb_001141_410 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 2 |
Hb_002769_020 |
0.0931193494 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 3 |
Hb_000203_230 |
0.1375127218 |
- |
- |
PREDICTED: probable inactive dual specificity protein phosphatase-like At4g18593 [Jatropha curcas] |
| 4 |
Hb_004375_130 |
0.1399022937 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 5 |
Hb_001488_340 |
0.1443942315 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 6 |
Hb_000700_150 |
0.1589288518 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_074449_010 |
0.1592991122 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 8 |
Hb_182516_010 |
0.1665117162 |
- |
- |
PREDICTED: uncharacterized protein LOC105641595 [Jatropha curcas] |
| 9 |
Hb_002559_040 |
0.1711507853 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 5 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000270_870 |
0.1736539308 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 11 |
Hb_000175_590 |
0.1767761227 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_012098_150 |
0.178677623 |
- |
- |
ARC5 protein [Manihot esculenta] |
| 13 |
Hb_000031_280 |
0.1790863364 |
- |
- |
- |
| 14 |
Hb_000270_160 |
0.179767261 |
- |
- |
PREDICTED: uncharacterized protein LOC102625351 [Citrus sinensis] |
| 15 |
Hb_000566_100 |
0.180083521 |
- |
- |
hypothetical protein JCGZ_17993 [Jatropha curcas] |
| 16 |
Hb_001396_280 |
0.1822338509 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 17 |
Hb_002681_170 |
0.1826931419 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 18 |
Hb_000920_030 |
0.1838928106 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_003376_050 |
0.1846338553 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 20 |
Hb_000172_540 |
0.1858472151 |
- |
- |
hypothetical protein POPTR_0005s08350g [Populus trichocarpa] |