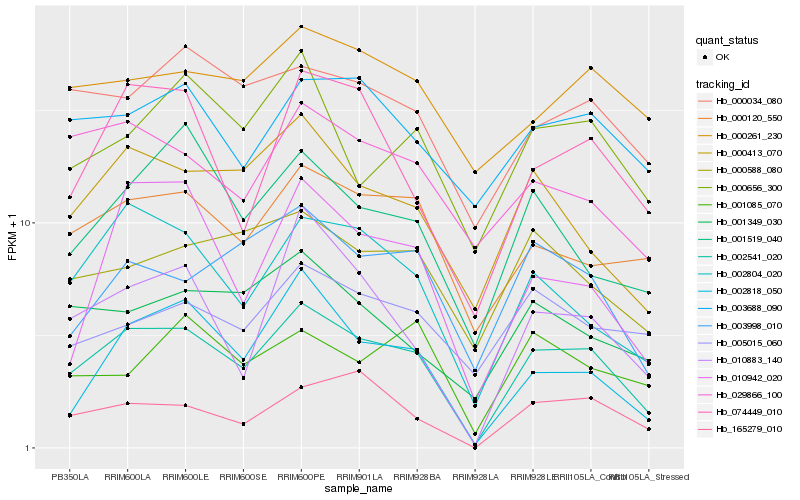
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002541_020 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 2 |
Hb_002804_020 |
0.1299410762 |
- |
- |
epsin N-terminal homology domain-containing family protein [Populus trichocarpa] |
| 3 |
Hb_000413_070 |
0.1446277884 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_074449_010 |
0.1465583932 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 5 |
Hb_000120_550 |
0.1472420832 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 6 |
Hb_165279_010 |
0.1481221614 |
- |
- |
Kiwellin, putative [Ricinus communis] |
| 7 |
Hb_002818_050 |
0.1515101832 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 8 |
Hb_010883_140 |
0.1539641013 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_003998_010 |
0.1581518921 |
- |
- |
PREDICTED: protein BRASSINOSTEROID INSENSITIVE 1 [Jatropha curcas] |
| 10 |
Hb_001519_040 |
0.1590338819 |
- |
- |
PREDICTED: uncharacterized protein LOC105636007 [Jatropha curcas] |
| 11 |
Hb_000034_080 |
0.1593616637 |
- |
- |
PREDICTED: MADS-box protein AGL24-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000656_300 |
0.1597375275 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 7 member B4-like [Populus euphratica] |
| 13 |
Hb_001349_030 |
0.1614977031 |
- |
- |
PREDICTED: uncharacterized protein LOC105641975 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000588_080 |
0.1629069771 |
- |
- |
PREDICTED: uncharacterized protein LOC105638459 [Jatropha curcas] |
| 15 |
Hb_029866_100 |
0.1629479813 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 16 |
Hb_003688_090 |
0.163769937 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 17 |
Hb_010942_020 |
0.1638011048 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000261_230 |
0.1718083195 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |
| 19 |
Hb_005015_060 |
0.1728706242 |
- |
- |
PREDICTED: developmentally-regulated G-protein 3 [Jatropha curcas] |
| 20 |
Hb_001085_070 |
0.1742253174 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |