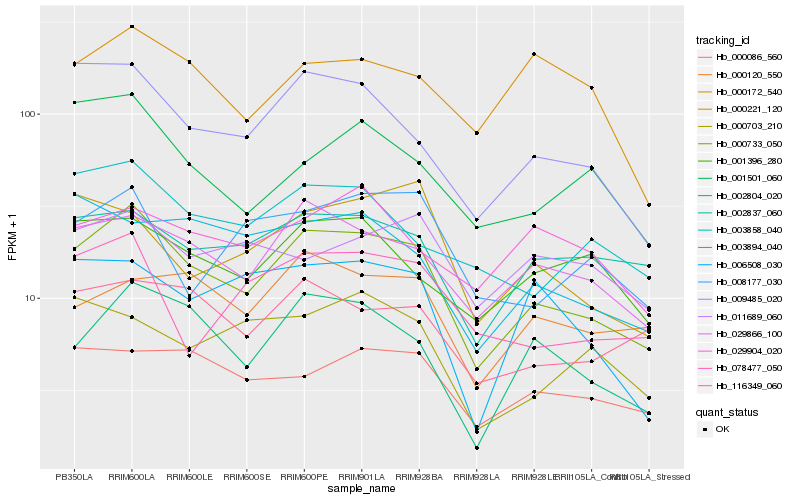
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000733_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646910 isoform X2 [Jatropha curcas] |
| 2 |
Hb_029866_100 |
0.1038073985 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 3 |
Hb_009485_020 |
0.1148028714 |
- |
- |
PREDICTED: membrane steroid-binding protein 2-like [Jatropha curcas] |
| 4 |
Hb_002804_020 |
0.1153729892 |
- |
- |
epsin N-terminal homology domain-containing family protein [Populus trichocarpa] |
| 5 |
Hb_002837_060 |
0.1313612413 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Nelumbo nucifera] |
| 6 |
Hb_116349_060 |
0.1355082634 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Brassica rapa] |
| 7 |
Hb_003858_040 |
0.1359637997 |
- |
- |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 8 |
Hb_001396_280 |
0.1362712787 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 9 |
Hb_000086_560 |
0.1371725109 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000120_550 |
0.1399786111 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 11 |
Hb_078477_050 |
0.1404046738 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105648332 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001501_060 |
0.1428087026 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000221_120 |
0.1453679051 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003894_040 |
0.1477254182 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 15 |
Hb_008177_030 |
0.1479207611 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_006508_030 |
0.1503486087 |
- |
- |
PREDICTED: CDPK-related kinase 1-like [Jatropha curcas] |
| 17 |
Hb_000172_540 |
0.1530297133 |
- |
- |
hypothetical protein POPTR_0005s08350g [Populus trichocarpa] |
| 18 |
Hb_000703_210 |
0.1541746988 |
- |
- |
Inactive protein kinase [Gossypium arboreum] |
| 19 |
Hb_011689_060 |
0.154471298 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 20 |
Hb_029904_020 |
0.1544784176 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |