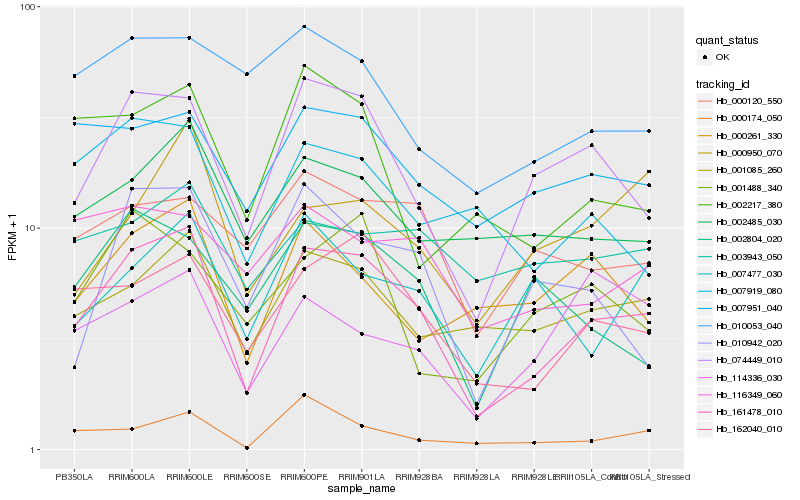
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_161478_010 |
0.0 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_074449_010 |
0.1401496695 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 3 |
Hb_162040_010 |
0.1466500511 |
- |
- |
- |
| 4 |
Hb_001085_260 |
0.1699522891 |
- |
- |
PREDICTED: uncharacterized protein LOC105633184 [Jatropha curcas] |
| 5 |
Hb_002485_030 |
0.1825549633 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 6 |
Hb_007951_040 |
0.1826975278 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 7 |
Hb_007477_030 |
0.1906921569 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |
| 8 |
Hb_010942_020 |
0.1907714051 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000120_550 |
0.19151785 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 10 |
Hb_002804_020 |
0.1919170349 |
- |
- |
epsin N-terminal homology domain-containing family protein [Populus trichocarpa] |
| 11 |
Hb_007919_080 |
0.1933189312 |
- |
- |
PREDICTED: uncharacterized protein At1g76660 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000261_330 |
0.1943308307 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Amborella trichopoda] |
| 13 |
Hb_116349_060 |
0.1949639279 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Brassica rapa] |
| 14 |
Hb_002217_380 |
0.1952503697 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 15 |
Hb_000174_050 |
0.1960780419 |
- |
- |
PREDICTED: gamma-tubulin complex component 2 [Populus euphratica] |
| 16 |
Hb_000950_070 |
0.1976815714 |
- |
- |
PREDICTED: uncharacterized protein LOC105645259 isoform X2 [Jatropha curcas] |
| 17 |
Hb_010053_040 |
0.1998641297 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 18 |
Hb_001488_340 |
0.2006346805 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 19 |
Hb_003943_050 |
0.2009350212 |
- |
- |
phosphoglucomutase, putative [Ricinus communis] |
| 20 |
Hb_114336_030 |
0.2019461684 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |