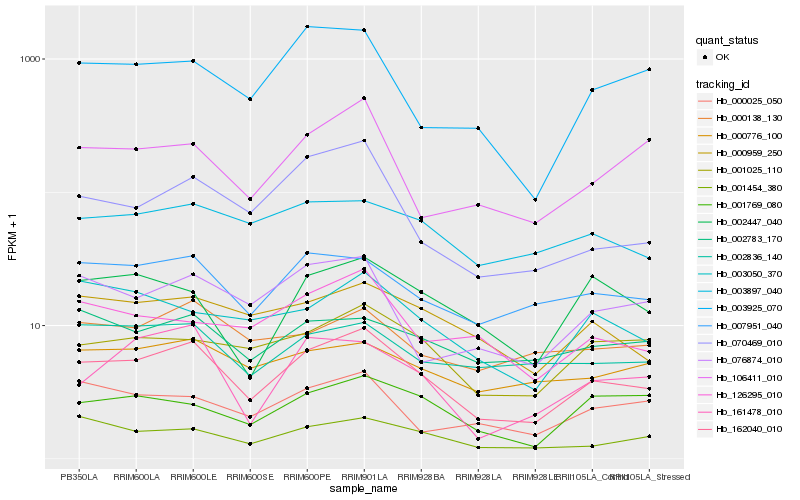
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_162040_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_007951_040 |
0.128044474 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 3 |
Hb_001025_110 |
0.1334456899 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001454_380 |
0.1349074321 |
- |
- |
PREDICTED: Fanconi anemia group I protein [Jatropha curcas] |
| 5 |
Hb_000959_250 |
0.1373495261 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase large subunit [Jatropha curcas] |
| 6 |
Hb_070469_010 |
0.1374510332 |
- |
- |
hypothetical protein POPTR_0002s21850g [Populus trichocarpa] |
| 7 |
Hb_003050_370 |
0.1429161784 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 8 |
Hb_076874_010 |
0.1442146701 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |
| 9 |
Hb_003897_040 |
0.1459746884 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 10 |
Hb_003925_070 |
0.1461220531 |
- |
- |
ATOZI1, putative [Ricinus communis] |
| 11 |
Hb_161478_010 |
0.1466500511 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_002836_140 |
0.1466930118 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 13 |
Hb_000776_100 |
0.1471519618 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |
| 14 |
Hb_001769_080 |
0.1478079858 |
- |
- |
hypothetical protein SORBIDRAFT_10g023200 [Sorghum bicolor] |
| 15 |
Hb_002447_040 |
0.1483164058 |
- |
- |
Cycloartenol-C-24-methyltransferase -like protein [Gossypium arboreum] |
| 16 |
Hb_126295_010 |
0.1486232184 |
- |
- |
PREDICTED: protein SEH1 [Jatropha curcas] |
| 17 |
Hb_000025_050 |
0.1490927626 |
- |
- |
hypothetical protein POPTR_0003s13040g [Populus trichocarpa] |
| 18 |
Hb_000138_130 |
0.1495683264 |
- |
- |
hypothetical protein JCGZ_25540 [Jatropha curcas] |
| 19 |
Hb_002783_170 |
0.1499042234 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X2 [Jatropha curcas] |
| 20 |
Hb_106411_010 |
0.1501576781 |
- |
- |
Ribosomal protein S25 family protein [Theobroma cacao] |