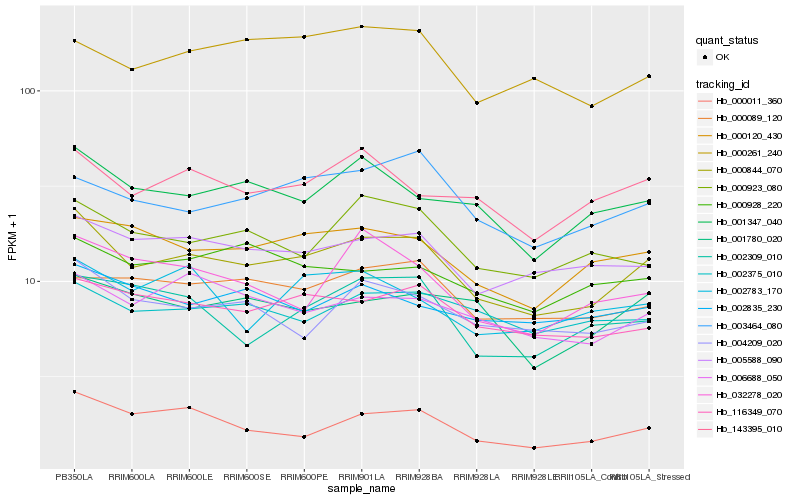
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000844_070 |
0.0 |
- |
- |
hypothetical protein POPTR_0014s03640g [Populus trichocarpa] |
| 2 |
Hb_000011_360 |
0.0873983016 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit alpha-like [Fragaria vesca subsp. vesca] |
| 3 |
Hb_005588_090 |
0.0887732835 |
- |
- |
hypothetical protein CICLE_v10003480mg [Citrus clementina] |
| 4 |
Hb_000120_430 |
0.0889952964 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 5 |
Hb_116349_070 |
0.08910769 |
- |
- |
hypothetical protein JCGZ_21753 [Jatropha curcas] |
| 6 |
Hb_001347_040 |
0.0904235442 |
- |
- |
PREDICTED: uncharacterized protein LOC105633920 [Jatropha curcas] |
| 7 |
Hb_004209_020 |
0.0929940964 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 51-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_143395_010 |
0.0944155842 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 9 |
Hb_006688_050 |
0.0950926545 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX15 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000923_080 |
0.0968333351 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 11 |
Hb_003464_080 |
0.0973517897 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 12 |
Hb_002309_010 |
0.1015352326 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 13 |
Hb_001780_020 |
0.1025425774 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000089_120 |
0.1038697899 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 15 |
Hb_000261_240 |
0.1039273861 |
- |
- |
RecName: Full=Elongation factor 1-alpha; Short=EF-1-alpha [Manihot esculenta] |
| 16 |
Hb_032278_020 |
0.1040907064 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 16 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002375_010 |
0.1045205507 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002835_230 |
0.1049680856 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000928_220 |
0.1059974933 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 20 |
Hb_002783_170 |
0.1070255385 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X2 [Jatropha curcas] |