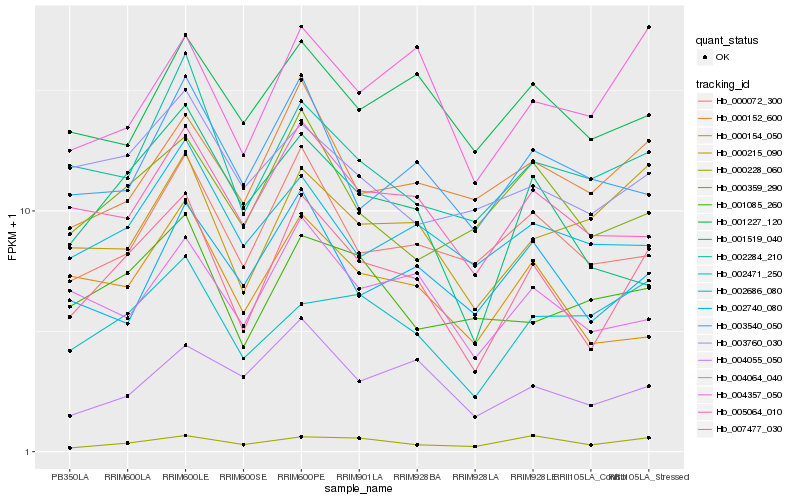
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007477_030 |
0.0 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |
| 2 |
Hb_005064_010 |
0.1351438623 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 3 |
Hb_002284_210 |
0.1413047484 |
- |
- |
PREDICTED: protein canopy-1 [Jatropha curcas] |
| 4 |
Hb_004064_040 |
0.1422390686 |
- |
- |
PREDICTED: Down syndrome critical region protein 3 homolog isoform X3 [Jatropha curcas] |
| 5 |
Hb_000072_300 |
0.1426624722 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Jatropha curcas] |
| 6 |
Hb_000154_050 |
0.1444922435 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 7 |
Hb_000215_090 |
0.1492466239 |
- |
- |
cytochrome b5 domain-containing family protein [Populus trichocarpa] |
| 8 |
Hb_000152_600 |
0.1502063408 |
- |
- |
PREDICTED: protein jagunal homolog 1 [Jatropha curcas] |
| 9 |
Hb_000359_290 |
0.1509710553 |
- |
- |
Triose phosphate/phosphate translocator, non-green plastid, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_001519_040 |
0.1522842558 |
- |
- |
PREDICTED: uncharacterized protein LOC105636007 [Jatropha curcas] |
| 11 |
Hb_002740_080 |
0.1523940801 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 12 |
Hb_004357_050 |
0.1526371393 |
- |
- |
PREDICTED: histone deacetylase HDT1-like [Jatropha curcas] |
| 13 |
Hb_001085_260 |
0.1545275571 |
- |
- |
PREDICTED: uncharacterized protein LOC105633184 [Jatropha curcas] |
| 14 |
Hb_004055_050 |
0.1548509335 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 15 |
Hb_002471_250 |
0.1555285661 |
- |
- |
Tapt1/CMV receptor, putative isoform 2 [Theobroma cacao] |
| 16 |
Hb_002686_080 |
0.1562225503 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein ING2-like [Jatropha curcas] |
| 17 |
Hb_000228_060 |
0.1567446833 |
- |
- |
hypothetical protein JCGZ_21532 [Jatropha curcas] |
| 18 |
Hb_003760_030 |
0.1570536522 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 19 |
Hb_003540_050 |
0.1575116637 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 20 |
Hb_001227_120 |
0.1589247453 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |