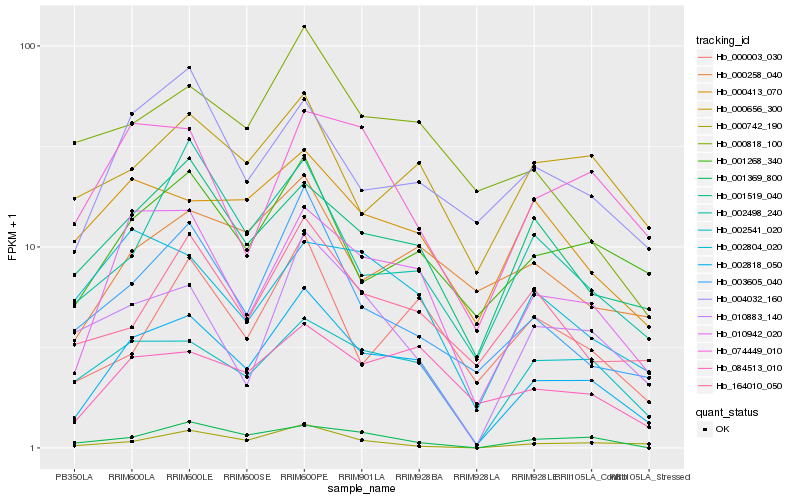
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002818_050 |
0.0 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 2 |
Hb_010942_020 |
0.1110864348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002541_020 |
0.1515101832 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 4 |
Hb_084513_010 |
0.1684302838 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 5 [Jatropha curcas] |
| 5 |
Hb_001519_040 |
0.1702683828 |
- |
- |
PREDICTED: uncharacterized protein LOC105636007 [Jatropha curcas] |
| 6 |
Hb_003605_040 |
0.1725728712 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001268_340 |
0.1731536483 |
- |
- |
Actin, putative [Ricinus communis] |
| 8 |
Hb_010883_140 |
0.1766766754 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_001369_800 |
0.1804745807 |
- |
- |
hypothetical protein B456_011G005000 [Gossypium raimondii] |
| 10 |
Hb_000742_190 |
0.1821799978 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_002498_240 |
0.1847945867 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_164010_050 |
0.1864069446 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |
| 13 |
Hb_004032_160 |
0.1873858045 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 14 |
Hb_000413_070 |
0.1903671104 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_074449_010 |
0.1906279281 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 16 |
Hb_002804_020 |
0.1916114162 |
- |
- |
epsin N-terminal homology domain-containing family protein [Populus trichocarpa] |
| 17 |
Hb_000258_040 |
0.1948770744 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 18 |
Hb_000003_030 |
0.1953375575 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_000656_300 |
0.1964336995 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 7 member B4-like [Populus euphratica] |
| 20 |
Hb_000818_100 |
0.1990798437 |
- |
- |
PREDICTED: delta(24)-sterol reductase [Jatropha curcas] |