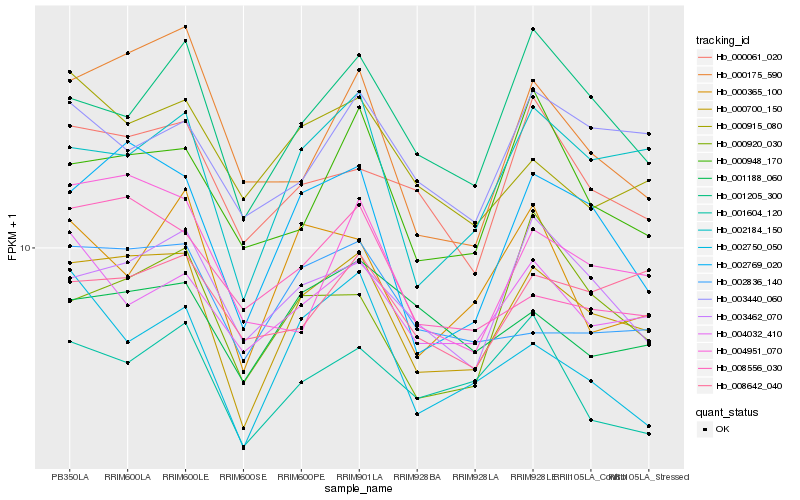
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000700_150 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002769_020 |
0.0850666926 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 3 |
Hb_002184_150 |
0.0884138164 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_001205_300 |
0.1080671958 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 5 |
Hb_003462_070 |
0.1103537507 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 6 |
Hb_004032_410 |
0.112404739 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000061_020 |
0.1136797526 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02 [Jatropha curcas] |
| 8 |
Hb_002836_140 |
0.1139499383 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 9 |
Hb_004951_070 |
0.1145579666 |
desease resistance |
Gene Name: AAA |
Stage III sporulation protein AA, putative [Ricinus communis] |
| 10 |
Hb_000915_080 |
0.1207543334 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 11 |
Hb_003440_060 |
0.1209453377 |
- |
- |
PREDICTED: translin-associated protein X isoform X1 [Jatropha curcas] |
| 12 |
Hb_000175_590 |
0.1209920274 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000365_100 |
0.1211543259 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000948_170 |
0.1216634447 |
- |
- |
hypothetical protein JCGZ_06749 [Jatropha curcas] |
| 15 |
Hb_008556_030 |
0.1218431051 |
- |
- |
PREDICTED: uncharacterized protein LOC105648021 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001604_120 |
0.1223394835 |
- |
- |
PREDICTED: hemK methyltransferase family member 1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000920_030 |
0.1225619858 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_008642_040 |
0.1227978252 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002750_050 |
0.1232435187 |
- |
- |
Chromosome-associated kinesin KLP1, putative [Ricinus communis] |
| 20 |
Hb_001188_060 |
0.1238236597 |
- |
- |
- |