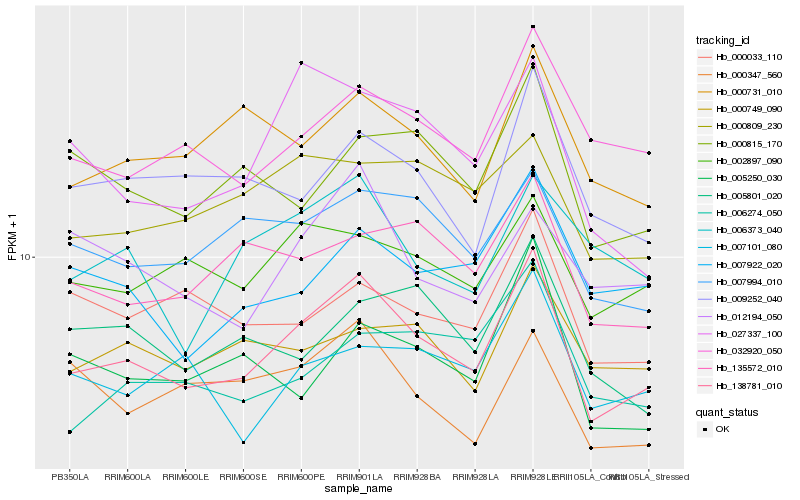
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_138781_010 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 2 |
Hb_032920_050 |
0.1180332483 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_007994_010 |
0.1213165923 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_135572_010 |
0.122047677 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 5 |
Hb_002897_090 |
0.1223883545 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000731_010 |
0.1245916936 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_009252_040 |
0.1246085066 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_027337_100 |
0.1257369612 |
- |
- |
Putative phagocytic receptor 1b [Gossypium arboreum] |
| 9 |
Hb_007922_020 |
0.1268938765 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 10 |
Hb_000815_170 |
0.1275089168 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |
| 11 |
Hb_000809_230 |
0.1282745238 |
- |
- |
PREDICTED: uncharacterized protein LOC105639681 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000749_090 |
0.1323332802 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_006274_050 |
0.1332350247 |
- |
- |
hypothetical protein VITISV_022618 [Vitis vinifera] |
| 14 |
Hb_005250_030 |
0.1334808417 |
- |
- |
PREDICTED: uncharacterized protein LOC105640177 [Jatropha curcas] |
| 15 |
Hb_005801_020 |
0.1338953489 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 16 |
Hb_007101_080 |
0.134452571 |
transcription factor |
TF Family: GNAT |
GCN5-related N-acetyltransferase family protein [Populus trichocarpa] |
| 17 |
Hb_006373_040 |
0.1349690274 |
- |
- |
PREDICTED: potassium transporter 11-like [Nicotiana tomentosiformis] |
| 18 |
Hb_000033_110 |
0.1351876783 |
- |
- |
hypothetical protein JCGZ_19253 [Jatropha curcas] |
| 19 |
Hb_000347_560 |
0.1370405013 |
- |
- |
- |
| 20 |
Hb_012194_050 |
0.1377933812 |
- |
- |
Aspartyl aminopeptidase, putative [Ricinus communis] |