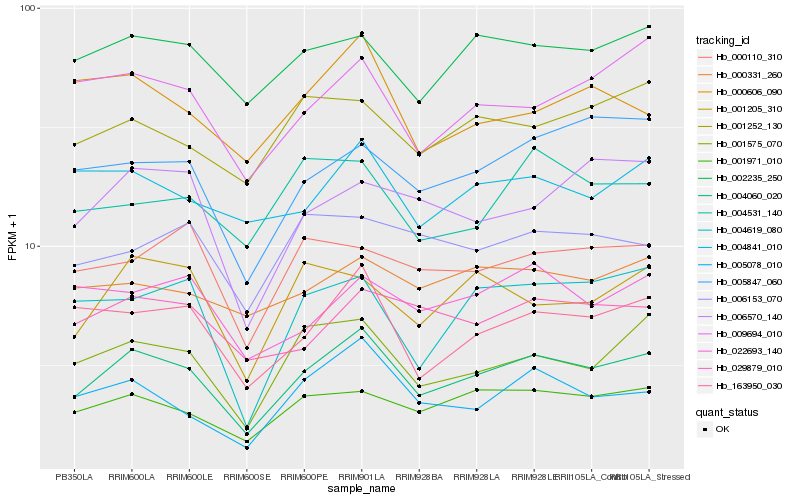
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004060_020 |
0.0 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_001971_010 |
0.08953069 |
- |
- |
F23N19.4 [Arabidopsis thaliana] |
| 3 |
Hb_001575_070 |
0.0904440241 |
- |
- |
PREDICTED: uncharacterized protein LOC105639885 [Jatropha curcas] |
| 4 |
Hb_163950_030 |
0.0926333106 |
- |
- |
PREDICTED: uncharacterized protein LOC105648509 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005078_010 |
0.0968325298 |
- |
- |
Derlin-2, putative [Ricinus communis] |
| 6 |
Hb_002235_250 |
0.1001439737 |
- |
- |
PREDICTED: cystathionine gamma-synthase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_009694_010 |
0.1062848715 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001252_130 |
0.1069420132 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 9 |
Hb_029879_010 |
0.1075266929 |
- |
- |
ribosomal pseudouridine synthase, putative [Ricinus communis] |
| 10 |
Hb_005847_060 |
0.1078106499 |
- |
- |
hypothetical protein POPTR_0006s02920g [Populus trichocarpa] |
| 11 |
Hb_001205_310 |
0.1085523879 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL4-like [Jatropha curcas] |
| 12 |
Hb_004841_010 |
0.1095553002 |
- |
- |
PREDICTED: 60S ribosomal protein L2, mitochondrial [Jatropha curcas] |
| 13 |
Hb_004619_080 |
0.1098281169 |
- |
- |
hypothetical protein PRUPE_ppa011369mg [Prunus persica] |
| 14 |
Hb_000331_260 |
0.1118381225 |
- |
- |
PREDICTED: WD repeat-containing protein 74 isoform X2 [Jatropha curcas] |
| 15 |
Hb_006570_140 |
0.1125954337 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 16 |
Hb_000110_310 |
0.1134162058 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 4 of pyruvate dehydrogenase complex |
Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_022693_140 |
0.1136333408 |
- |
- |
PREDICTED: uncharacterized protein LOC105632654 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004531_140 |
0.1147869614 |
- |
- |
PREDICTED: uncharacterized protein LOC105635023 [Jatropha curcas] |
| 19 |
Hb_006153_070 |
0.1148910624 |
- |
- |
3-oxoacyl-[acyl-carrier-protein] synthase 3 A, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000606_090 |
0.1152387278 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |