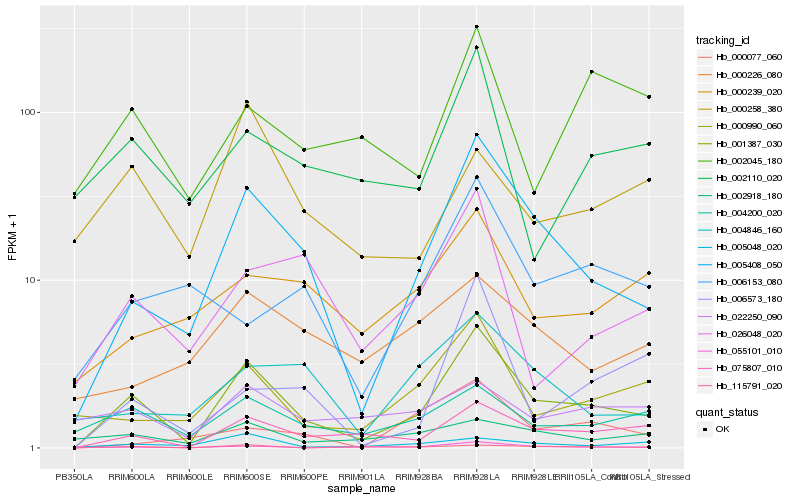
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001387_030 |
0.0 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0013s13920g [Populus trichocarpa] |
| 2 |
Hb_005408_050 |
0.1642038431 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK5 [Jatropha curcas] |
| 3 |
Hb_075807_010 |
0.2001803592 |
- |
- |
PREDICTED: uncharacterized protein LOC105631775 [Jatropha curcas] |
| 4 |
Hb_055101_010 |
0.2202833876 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_004200_020 |
0.2506452471 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 6 |
Hb_000990_060 |
0.2530192298 |
- |
- |
PREDICTED: protein EMBRYONIC FLOWER 1 [Jatropha curcas] |
| 7 |
Hb_115791_020 |
0.2640003591 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 8 |
Hb_005048_020 |
0.2647202758 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 9 |
Hb_026048_020 |
0.2773269942 |
- |
- |
PREDICTED: uncharacterized protein LOC105645738 [Jatropha curcas] |
| 10 |
Hb_004846_160 |
0.280294338 |
- |
- |
- |
| 11 |
Hb_002918_180 |
0.2812674345 |
- |
- |
hypothetical protein POPTR_0004s04740g [Populus trichocarpa] |
| 12 |
Hb_002110_020 |
0.2829926131 |
- |
- |
PREDICTED: transcription factor bHLH145 [Jatropha curcas] |
| 13 |
Hb_006573_180 |
0.2877471021 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1581940 [Ricinus communis] |
| 14 |
Hb_000239_020 |
0.2898369869 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g43190 [Jatropha curcas] |
| 15 |
Hb_022250_090 |
0.2927689165 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08070-like [Jatropha curcas] |
| 16 |
Hb_000077_060 |
0.2947186324 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH36-like [Jatropha curcas] |
| 17 |
Hb_002045_180 |
0.2952632142 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 18 |
Hb_000226_080 |
0.2968499086 |
- |
- |
prephenate dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_000258_380 |
0.3022238866 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP11-like [Jatropha curcas] |
| 20 |
Hb_006153_080 |
0.3034549837 |
- |
- |
flavin-containing monooxygenase-related family protein [Populus trichocarpa] |