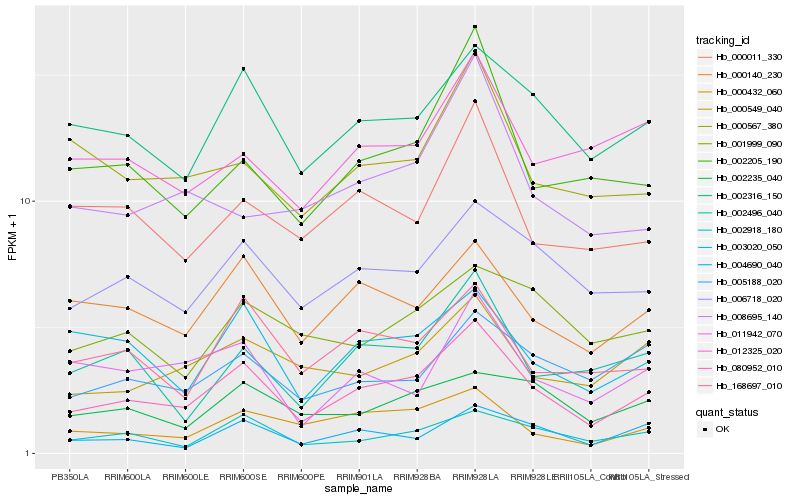
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_080952_010 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20770 [Jatropha curcas] |
| 2 |
Hb_002316_150 |
0.1231041746 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 3 |
Hb_006718_020 |
0.1242909041 |
- |
- |
PREDICTED: uncharacterized protein LOC105649330 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000140_230 |
0.1248520828 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61520, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_002918_180 |
0.126100639 |
- |
- |
hypothetical protein POPTR_0004s04740g [Populus trichocarpa] |
| 6 |
Hb_000432_060 |
0.1272045197 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_168697_010 |
0.1301781174 |
- |
- |
hypothetical protein JCGZ_16772 [Jatropha curcas] |
| 8 |
Hb_002205_190 |
0.1308197422 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 9 |
Hb_003020_050 |
0.1318065656 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000549_040 |
0.1330595298 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_005188_020 |
0.1370309176 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g02330 [Jatropha curcas] |
| 12 |
Hb_004690_040 |
0.1381566683 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71420 [Jatropha curcas] |
| 13 |
Hb_002235_040 |
0.1397169508 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230 [Jatropha curcas] |
| 14 |
Hb_002496_040 |
0.139936353 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03380, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000011_330 |
0.1404850732 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 16 |
Hb_000567_380 |
0.1430817961 |
- |
- |
ubiquitin specific protease 39 and snrnp assembly factor, putative [Ricinus communis] |
| 17 |
Hb_011942_070 |
0.1442637232 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g20230-like [Jatropha curcas] |
| 18 |
Hb_008695_140 |
0.1494732272 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 19 |
Hb_012325_020 |
0.1513581937 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein At1g05910 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001999_090 |
0.1520335076 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g29760, chloroplastic [Jatropha curcas] |