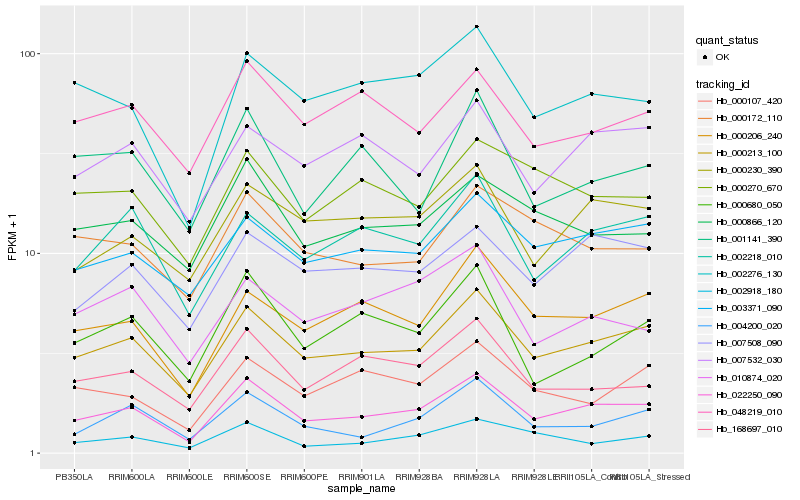
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022250_090 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08070-like [Jatropha curcas] |
| 2 |
Hb_000213_100 |
0.0603476412 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79540-like [Jatropha curcas] |
| 3 |
Hb_000206_240 |
0.1023733403 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002276_130 |
0.1042362771 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 5 |
Hb_002218_010 |
0.1043580203 |
transcription factor |
TF Family: Orphans |
ethylene receptor, putative [Ricinus communis] |
| 6 |
Hb_000230_390 |
0.1061934221 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880 [Jatropha curcas] |
| 7 |
Hb_003371_090 |
0.1078205453 |
- |
- |
PREDICTED: uncharacterized protein LOC105650336 [Jatropha curcas] |
| 8 |
Hb_007508_090 |
0.1086897532 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 9 |
Hb_168697_010 |
0.1136822748 |
- |
- |
hypothetical protein JCGZ_16772 [Jatropha curcas] |
| 10 |
Hb_004200_020 |
0.1141364795 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 11 |
Hb_000866_120 |
0.1159668796 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12 [Jatropha curcas] |
| 12 |
Hb_000680_050 |
0.1167256614 |
- |
- |
hypothetical protein JCGZ_24767 [Jatropha curcas] |
| 13 |
Hb_007532_030 |
0.118238927 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 14 |
Hb_000107_420 |
0.1215188405 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03880, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000172_110 |
0.1219003664 |
- |
- |
- |
| 16 |
Hb_048219_010 |
0.1238241807 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 17 |
Hb_000270_670 |
0.1241549665 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 18 |
Hb_002918_180 |
0.12459759 |
- |
- |
hypothetical protein POPTR_0004s04740g [Populus trichocarpa] |
| 19 |
Hb_010874_020 |
0.1250491664 |
- |
- |
PREDICTED: uncharacterized protein LOC105636745 [Jatropha curcas] |
| 20 |
Hb_001141_390 |
0.1257014218 |
- |
- |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |