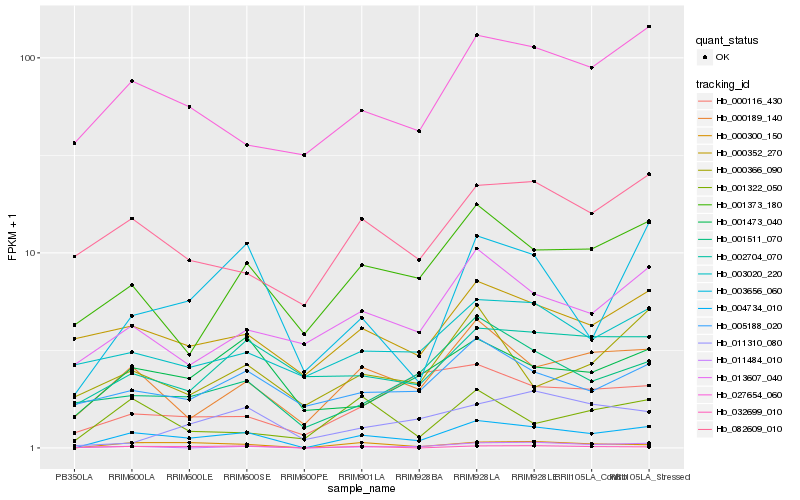
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004734_010 |
0.0 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000300_150 |
0.1657818618 |
- |
- |
Peroxidase 44 precursor, putative [Ricinus communis] |
| 3 |
Hb_000189_140 |
0.1975170265 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_003656_060 |
0.2155577849 |
- |
- |
monooxygenase, putative [Ricinus communis] |
| 5 |
Hb_001373_180 |
0.225381715 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001473_040 |
0.2275677818 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 7 |
Hb_001511_070 |
0.2310829482 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0010s19310g [Populus trichocarpa] |
| 8 |
Hb_027654_060 |
0.2343880669 |
- |
- |
RecName: Full=Probable pyridoxal 5'-phosphate synthase subunit PDX1; Short=PLP synthase subunit PDX1; AltName: Full=Ethylene-inducible protein HEVER [Hevea brasiliensis] |
| 9 |
Hb_013607_040 |
0.2373325902 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18520-like [Jatropha curcas] |
| 10 |
Hb_082609_010 |
0.2387449802 |
- |
- |
PREDICTED: uncharacterized protein LOC105644299 [Jatropha curcas] |
| 11 |
Hb_005188_020 |
0.2401814178 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g02330 [Jatropha curcas] |
| 12 |
Hb_002704_070 |
0.2410118769 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like [Jatropha curcas] |
| 13 |
Hb_000352_270 |
0.2416956635 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g26630, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_003020_220 |
0.2444655044 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000116_430 |
0.2457694433 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 1, mitochondrial-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001322_050 |
0.2463155952 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16835, mitochondrial [Jatropha curcas] |
| 17 |
Hb_032699_010 |
0.2488438269 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_011484_010 |
0.2512043245 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 19 |
Hb_000366_090 |
0.2515274606 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32430, mitochondrial [Jatropha curcas] |
| 20 |
Hb_011310_080 |
0.2519041858 |
- |
- |
hypothetical protein POPTR_0006s26840g [Populus trichocarpa] |