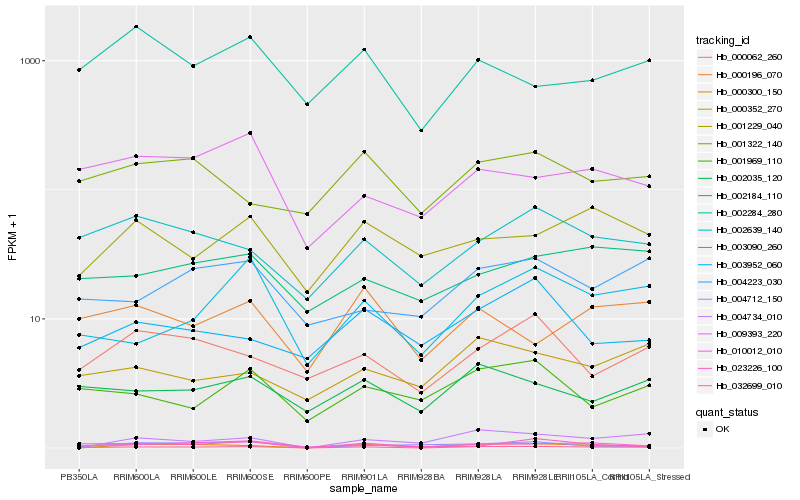
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032699_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_010012_010 |
0.2057448612 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000300_150 |
0.2107879751 |
- |
- |
Peroxidase 44 precursor, putative [Ricinus communis] |
| 4 |
Hb_003090_260 |
0.2339989393 |
- |
- |
hypothetical protein JCGZ_25097 [Jatropha curcas] |
| 5 |
Hb_002639_140 |
0.2416684623 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 5 [Jatropha curcas] |
| 6 |
Hb_004734_010 |
0.2488438269 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 7 |
Hb_023226_100 |
0.2509509837 |
- |
- |
PREDICTED: uncharacterized protein LOC104903172 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_000062_260 |
0.2521084628 |
- |
- |
PREDICTED: uncharacterized protein LOC105644756 [Jatropha curcas] |
| 9 |
Hb_002035_120 |
0.2521322451 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g14330 [Jatropha curcas] |
| 10 |
Hb_009393_220 |
0.2523624168 |
transcription factor |
TF Family: Tify |
JAZ11 [Hevea brasiliensis] |
| 11 |
Hb_001969_110 |
0.2609258282 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g15510, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004223_030 |
0.2612136049 |
- |
- |
PREDICTED: uncharacterized protein LOC105635347 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001322_140 |
0.2647429738 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001229_040 |
0.2653963033 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 15 |
Hb_002184_110 |
0.2659614206 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 [Jatropha curcas] |
| 16 |
Hb_000196_070 |
0.2673226577 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH80-like [Jatropha curcas] |
| 17 |
Hb_004712_150 |
0.2673774641 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_003952_060 |
0.2674113068 |
- |
- |
hypothetical protein POPTR_0016s11000g [Populus trichocarpa] |
| 19 |
Hb_000352_270 |
0.2676747727 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g26630, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_002284_280 |
0.2686278449 |
transcription factor |
TF Family: HMG |
high mobility group protein [Hevea brasiliensis] |