| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
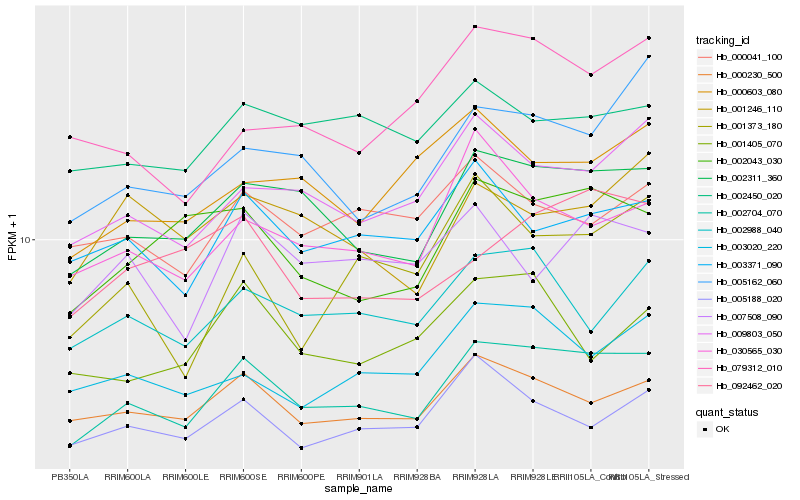
Hb_002704_070 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like [Jatropha curcas] |
| 2 |
Hb_002311_360 |
0.0769947663 |
- |
- |
PREDICTED: uncharacterized protein LOC105114273 [Populus euphratica] |
| 3 |
Hb_000230_500 |
0.1024415316 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g19020, mitochondrial [Jatropha curcas] |
| 4 |
Hb_009803_050 |
0.1065447086 |
- |
- |
PREDICTED: uncharacterized protein LOC105644460 [Jatropha curcas] |
| 5 |
Hb_002043_030 |
0.1091335374 |
- |
- |
hypothetical protein RCOM_1486170 [Ricinus communis] |
| 6 |
Hb_030565_030 |
0.1148262107 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1-like [Jatropha curcas] |
| 7 |
Hb_002988_040 |
0.1162869777 |
- |
- |
- |
| 8 |
Hb_003371_090 |
0.117909628 |
- |
- |
PREDICTED: uncharacterized protein LOC105650336 [Jatropha curcas] |
| 9 |
Hb_005162_060 |
0.1183001981 |
- |
- |
PREDICTED: putative septum site-determining protein minD homolog, chloroplastic [Jatropha curcas] |
| 10 |
Hb_003020_220 |
0.1188533125 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |
| 11 |
Hb_005188_020 |
0.1217005384 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g02330 [Jatropha curcas] |
| 12 |
Hb_092462_020 |
0.1250547467 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_001373_180 |
0.1268328733 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000603_080 |
0.1273271826 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33170 [Vitis vinifera] |
| 15 |
Hb_002450_020 |
0.128095175 |
- |
- |
PREDICTED: probable transcriptional regulator SLK2 [Jatropha curcas] |
| 16 |
Hb_079312_010 |
0.1282204423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007508_090 |
0.1285647221 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 18 |
Hb_001246_110 |
0.1294224151 |
- |
- |
PREDICTED: probable purine permease 11 [Jatropha curcas] |
| 19 |
Hb_000041_100 |
0.1294687742 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 41 [Jatropha curcas] |
| 20 |
Hb_001405_070 |
0.1301010127 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g23020 [Jatropha curcas] |