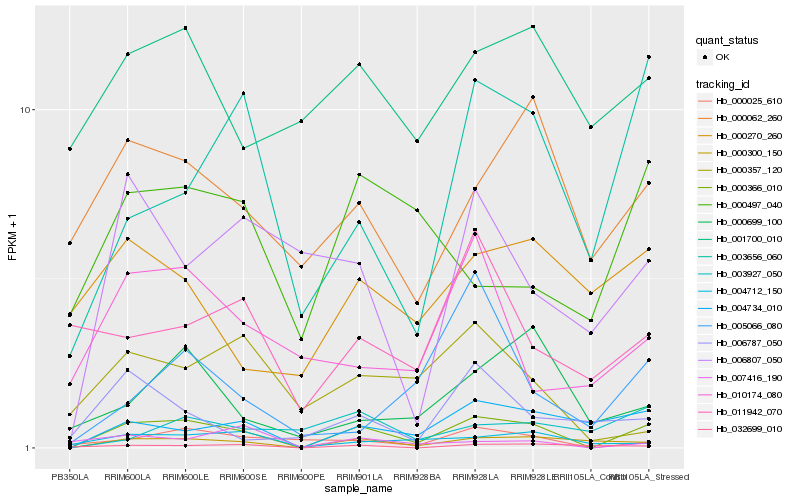
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000366_010 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08070-like, partial [Pyrus x bretschneideri] |
| 2 |
Hb_000300_150 |
0.2430322782 |
- |
- |
Peroxidase 44 precursor, putative [Ricinus communis] |
| 3 |
Hb_004734_010 |
0.2675564091 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000025_610 |
0.2862189732 |
- |
- |
annexin, putative [Ricinus communis] |
| 5 |
Hb_003656_060 |
0.2945438661 |
- |
- |
monooxygenase, putative [Ricinus communis] |
| 6 |
Hb_004712_150 |
0.3088025047 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_000062_260 |
0.3166088865 |
- |
- |
PREDICTED: uncharacterized protein LOC105644756 [Jatropha curcas] |
| 8 |
Hb_000699_100 |
0.3188495187 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g01510, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_032699_010 |
0.3202335818 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 10 |
Hb_006807_050 |
0.3226831412 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPH8A, putative [Ricinus communis] |
| 11 |
Hb_005066_080 |
0.3346444514 |
- |
- |
hypothetical protein PRUPE_ppa025081mg [Prunus persica] |
| 12 |
Hb_000357_120 |
0.3355554429 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like [Jatropha curcas] |
| 13 |
Hb_010174_080 |
0.3360543941 |
- |
- |
- |
| 14 |
Hb_001700_010 |
0.3361392556 |
- |
- |
PREDICTED: GTP-binding protein At2g22870 [Jatropha curcas] |
| 15 |
Hb_006787_050 |
0.3366126886 |
- |
- |
hypothetical protein JCGZ_23988 [Jatropha curcas] |
| 16 |
Hb_011942_070 |
0.3386309096 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g20230-like [Jatropha curcas] |
| 17 |
Hb_007416_190 |
0.3394399043 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Populus euphratica] |
| 18 |
Hb_003927_050 |
0.340708492 |
- |
- |
PREDICTED: uncharacterized protein LOC105629028 [Jatropha curcas] |
| 19 |
Hb_000497_040 |
0.3408735014 |
- |
- |
BnaC09g09960D [Brassica napus] |
| 20 |
Hb_000270_260 |
0.3420367765 |
- |
- |
PREDICTED: uncharacterized protein LOC105640466 [Jatropha curcas] |