| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
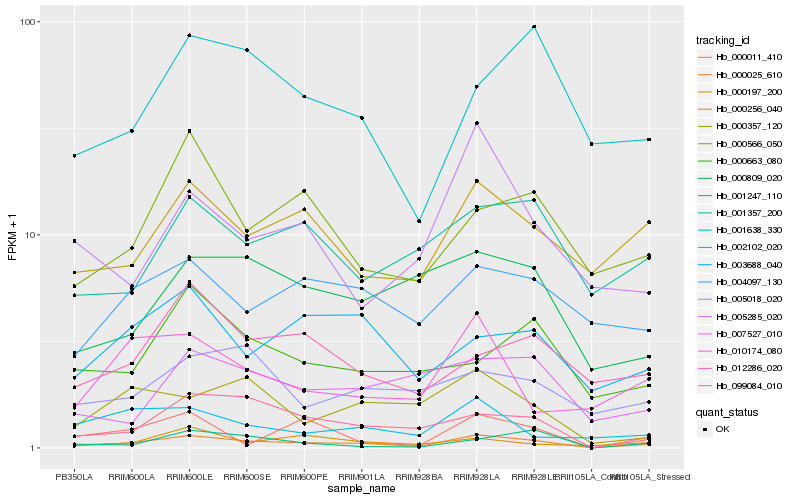
Hb_000025_610 |
0.0 |
- |
- |
annexin, putative [Ricinus communis] |
| 2 |
Hb_099084_010 |
0.1893143751 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 3 |
Hb_000256_040 |
0.2185972704 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000566_050 |
0.2250562608 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 5 |
Hb_000809_020 |
0.2287539821 |
desease resistance |
Gene Name: RPW8 |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_001638_330 |
0.2307385174 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003688_040 |
0.2313767804 |
- |
- |
- |
| 8 |
Hb_012286_020 |
0.2320836593 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 9 |
Hb_001357_200 |
0.2334220206 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 10 |
Hb_004097_130 |
0.2345652451 |
- |
- |
Inositol-tetrakisphosphate 1-kinase, putative [Ricinus communis] |
| 11 |
Hb_000357_120 |
0.2352642617 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like [Jatropha curcas] |
| 12 |
Hb_010174_080 |
0.2352873581 |
- |
- |
- |
| 13 |
Hb_007527_010 |
0.2365182475 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001247_110 |
0.2377490225 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 15 |
Hb_005018_020 |
0.2404548745 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g07740, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000011_410 |
0.2406637641 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000197_200 |
0.2412175655 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 18 |
Hb_005285_020 |
0.2415549533 |
- |
- |
- |
| 19 |
Hb_000663_080 |
0.2424790321 |
transcription factor |
TF Family: HB |
Homeobox protein HAT3.1, putative [Ricinus communis] |
| 20 |
Hb_002102_020 |
0.2436517708 |
- |
- |
PREDICTED: lysine-specific histone demethylase 1 homolog 1 [Jatropha curcas] |